Understanding the Central Dogma of Molecular Biology - DNA, Chromosomes, and Gene Regulation
Delve into the intricate world of molecular biology as we explore the central dogma, from DNA structure and replication to gene regulation and chromosomal organization in eukaryotic cells. Discover the fundamental principles governing genetic information flow and genome evolution, offering insights into the foundation of life itself.
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author. Download presentation by click this link. If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
E N D
Presentation Transcript
Central Dogma of Molecular Biology Cell Bio Ch 5- 10 Sattvik Basarkod
Table of Contents Chapter 5: DNA and Chromosomes Chapter 6: DNA Replication Chapter 7: Transcription/ Translation Chapter 8: Regulation of Genes Chapter 9: Evolution of Genomes Chapter 10: Analysis of Genes
Chapter 5: DNA and Chromosomes DNA Mitochondria with solid fill Police male outline DNA Structure DNA STRUCTURE Eukaryotic Chromosomes EUKARYOTIC CHROMOSOMES Regulation of Chromosomes REGULATION OF CHROMOSOMES Regulation of Chromosomes Back to Table of Contents
DNA Structure - Backbone of DNA consists of deoxyribose sugar and phosphate group The phosphate group is at 5 end and the sugar group is at 3 end Four Types of Nucleic Acids: A and T(purines) and C and G(pyrimidines) are hydrogen bonded Purines have 2 rings; Pyrimidines have 3 rings. A and T form 2 bonds; C and G form 3. Nucleotides are held together by phosphodiester bonds- pyrophosphorylation provides energy - Antiparallel strands of DNA create double helix. Back to Chapter 5 From Textbook pg. 176
Eukaryotic Chromosomes - Prokaryotes have circular chromosomes, while eukaryotes have linear chromosomes - Nucleosomes are basic repeating units of eukaryotic chromosomes(chromatin) - Histone octamer and DNA complex create chromatin complex - Nucleosome < Chromatin < Chromatin Fibers < Loops of chromatin fibers < mitotic chromosomes Positive charges on histone amino acids strongly interact with the negatively charged DNA backbone Chromosomes contain genomes, which have coding areas called genes. Back to Chapter 5 From textbook pg. 186
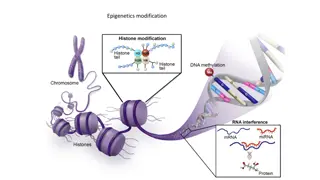
Regulation of Chromosomes - Changes in Nucleosomes Chromatin Remodeling complexes roll histones around different DNA areas Histone Modifying enzymes add/remove phosphate/ acetyl/ methyl group to modify histones. This can result in de-condensation or condensation of DNA - Heterochromatin is highly condensed and not expressed( in telomeres and centromeres) Heterochromatin will continue to spread until a barrier DNA sequence is found. Main job of heterochromatin is to silence genes- to inhibit their gene expression. - Euchromatin is less condensed and is expressed (coding genes that form proteins) - One random X chromosome is inactivated in females and seen as a Barr Body: X inactivation. From textbook pg. 190 - Back to Chapter 5
Chapter 6: DNA Replication Fork In Road with solid fill DNA Checklist outline Replication Forks Replication Forks DNA Synthesis DNA Synthesis Telomerase Telomerase Back to Table of Contents
Replication Forks - DNA replication is done through complementary base pairing of template and new strand: semiconservative model. - Eukaryotic DNA has many replication origins, one centromere, and two telomers. Prokaryotes only have one origin of replication - Replication forks are at the 3 end of the template strand. Nucleotides are added to the 3 end of the template strand. - Each replication origin has two forks, opening each strand of the DNA. One strand is leading strand(5 - 3 end) and the other is lagging strand(3 -5 end) - Bidirectional replication is present in both eukaryotes and prokaryotes. Back to Chapter 6 From textbook pg. 201
DNA Synthesis - Steps of DNA Synthesis - Initiator proteins break hydrogen bonds of the double helix DNA. DNA helicase unzips DNA as well. - Primase/ RNA polymerase is used to initiate the process of DNA replication. - DNA polymerase 3 synthesizes new nucleotides to the template strand. The base pairs bind, and backbone is connected by energy from pyrophosphates. - Leading Strand is replicated continuously from 5 to 3 end of new strand. DNA Polymerase 3 can only work in one direction; thus, the lagging strand forms Okazaki fragments. - Nuclease cuts the DNA into Okazaki fragments; DNA polymerase adds DNA primers at new 3 end and ligase connects all the fragments. - SSB(Single Stranded Binding Proteins prevent base pair of the two template strands. - DNA topoisomerase slightly winds the DNA to relive tension in the replication fork From textbook pg. 211 - Back to Chapter 6
Telomerase - Leading strand can be continuously replicated all the way up to the end; however, the lagging strand shortens at the end. - In bacteria, this problem is solved due to their circular chromosomes. - In eukaryotes, this problem is solved by using telomerase which adds telomers to the end of the lagging strand template, allowing final Okazaki fragment to form. - Telomerase binds to the template strand by using its RNA template - Telomeres are shortened with age and other autoimmune disorders. From textbook pg. 214 DNA polymerase is self correcting- matches pairs at a high rate- if it makes a mistake, it proofreads previous sequence Back to Chapter 6
Chapter 7: Transcription/ Translation Two Men with solid fill DNA with solid fill RNA vs DNA RNA vs DNA Transcription Transcription Translation Translation Protein Regulation Protein Regulation Transcription Table Transcription Table Translation Table Translation Table Back to Table of Contents
RNA VS DNA From textbook pg. 279 RNA Both DNA Contains two OH group on sugar: ribose. Less stable Base Pairs: A, C, G Contains one OH group on sugar: deoxyribose. More stable Contains Uracil(U) not Thymine(T) Hydrogen bonds between the base pairs. Contains Thymine(T) not Uracil(U) Types of RNA: mRNA, tRNA, rRNA siRNA, miRNA, Phosphodiester bonds between the nucleotides. Types of DNA: cDNA, Used in Transcription, Translation Similar backbone except the sugar(contains OH) Used in Replication, Transcription Back to Chapter 7 Singled stranded Both are synthesized 5 to 3 on new strand Double Stranded (can be double stranded for viruses)
Transcription - Transcription is a process where double stranded DNA is converted in single stranded mRNA. RNA polymerase unwinds and winds double helix to connect ribonucleic acids to the deoxynucleic acids. Strands of DNA serve as templates to form mRNA. The mRNA does not remain hydrogen bonded to the DNA strand, as RNA poly II rewinds the DNA as it moves forward. Like DNA Poly, RNA poly II also uses the pyrophosphate energy to create phosphodiester bonds between the nucleotides to form the mRNA. A promotor site signals RNA poly II to start transcription. RNA polymerase II can only synthesize in 5 -3 direction so the it must use 3 -5 DNA as template. - Prokaryotic Transcription uses Sigma factor to help RNA poly II find the promotor. The promotor is not the start site, but it signals for it. - Eukaryotic Transcription requires many general transcription factors to assemble on the promotor to start transcription. By using mediators, TF2D, TF2H, TATA box, and other regulators, the RNA polymerase starts transcription. Now the premature mRNA must become stable enough to leave the nucleus for translation. For this, there is a 5 cap and a poly A tail added. The mRNA is also spliced by spliceosomes, which recognize mRNA introns by snRNA and remove them. This then leaves only exons, which are converted into protein in translation Back to Chapter 7
Transcription Table Eukaryotes Both Prokaryotes Happens in the nucleus Both contain promotor regions Happens in the cytoplasm Use RNA poly. I,II, III TATA box Use RNA poly I Use of transcription factors, mediators, enhancers and other protein complexes Both have repressors/ activators regulating gene expression Use of Sigma factor to initiate transcription 5 cap, remove exons, splice introns, add poly A tail to create mRNA Both degrade unused mRNA and recycle base pairs No 5 cap, splicing or poly A tail added. No mRNA Leaves nucleus for translation Both create one stranded RNA in transcription Translation and Transcription happen simultaneously Back to Chapter 7
Translation - Once eukaryotic mRNA arrives at cytosol, outside of nucleus, it is held together by two ribosomal units. The small subunit holds the mRNA, and the large subunit holds the tRNA which brings anticodons. A codon on mRNA is 3 nucleic acid sequence which codes for a specific amino acid. The start codon for translation is AUG, methionine amino acid. - The tRNA attaches to its specific amino acid by using aminoacyl-tRNA synthetase. The coupling of ATP hydrolysis allows strong energy for the amino acid to bind to the tRNA. The ribosome is a ribozyme which acts as a protein with rRNA recognizing the codons on the mRNA. Polyribosomes speed up the process of translation. - There are three sites of the ribosome: E, P, A. The first tRNA arrives at the P site, but after that all incoming tRNAs arrive at the A site. During the process of elongation, the tRNA amino acids create a peptide chain. The amino acids are combined by connecting the amino group and the carboxyl group. As the large subunit moves from 5 to 3 , previously used tRNA become uncharged and leave at the E site. This process is continued until there is a release factor. The release factor adds water which helps terminate the peptide chain from the tRNA. All enzymes are then recycled for another protein translation. Back to Chapter 7
Translation Table Eukaryotes Both Prokaryotes Uses 5 cap, translation initiation factors, and initiator tRNA Both have initiation, elongation, and termination Ribosome in prokaryotes directly binds to AUG at mRNA Regulation by using translation initiation factors Contain release factors Regulation by using 6 nucleotide ribosome binding sequence 80s ribosomes rRNA ribosomal units 70s ribosomes Monocistronic mRNA: code for only one protein E,P,A sites Polycistronic: Code for several different proteins on the same mRNA Back to Chapter 7
Protein Regulation - Proteins in body can be regulated by proteases, which hydrolyze peptide bond between the amino acids. This breaks down the protein molecule. - Proteosomes degrade set of proteins by binding to them. Covalent attachment of small molecules such as ubiquitin, signal to the proteosomes to bind to the protein and break it down. Back to Chapter 7
Chapter 8: Regulation of Genes DNA Microscope Pencil Document Gene Expression Gene Expression Transcription Regulation Transcription Regulation Specialized Cells Specialized Cells Post Transcriptional Control Post Transcriptional Control Transcription Regulation Post Transcriptional Control Back to Table of Contents
Gene Expression - Different cells have same DNA but different proteins and gene expression From textbook pg. 271 - Ways of regulating genes: Hormone signaling, RNA processing, RNA localization, rate of degradation of RNA/ protein, and protein turning on/ off - Major Controlling factor: Transcriptional changes. Back to Chapter 8
Transcription Regulation - Transcriptional regulators bind to DNA strand s major grooves in both prokaryotes and eukaryotes. - The genome s regulatory regions must be recognized by transcription regulators, which can either inhibit or activate gene expression. - Operons are regulatory units in bacteria. Tryp repressor allosterically binds to operator and sends positive feedback to stop making protein where there is enough tryptophan. It is inactive when there is no tryptophan. From textbook pg. 275 - Lac Operon is used to produce lactase protein. The operon has a Lac repressor and a CAP activator to regulate the expression. The lac repressor is active when there is no lactose, and the CAP activator is active when there is no glucose. When lac repressor is inactive and CAP activator is active, operon is on and the gene is expressed. This makes sense because in the presence of glucose, you need other type of carbohydrate to break down for energy. And the presence of lactose is necessary for lactase to work in the first place. Back to Chapter 8 * Histone modifications can cause chromatin remodeling, and this can also regulate gene expression.
Specialized Cells From textbook pg. 285 - Embryonic Stem Cells are totipotent/ call become any cell the body wants. - Cells differentiate based on their location and the proteins they their DNA codes for - Many regulatory steps can occur when a substrate binds to its receptor and signals other activators to turn on. - For example: Cortisol turns on several proteins to express a certain gene. - Master Transcription regulators can code for proteins for a whole organ - Epigenetics is the field of study that focuses on epigenomes caused by cell memory. DNA methylation of histone deacetylation can increase gene expression of respective DNA. These epigenome changes can be passed down to offspring or can last entire lifespan due to behavioral and genetic changes. An epigenetic inheritance research study by Daniel A Hackman Martha J. Farah shows that rats who did not get parental care had their genes altered by methylenation of histones. They were reserved and more stressed for their whole life. Back to Chapter 8
Post Transcriptional Control - miRNAs directly destruct mRNA by using RISC complex. RISC complex is a combination of miRNA and enzymes. The complex recognizes specific mRNA sequence and then breaks it down by nuclease enzyme. - siRNA are used in RNA interference, in which the body breaks down foreign substances such as viruses and bacteria. They can be paired up with RITS complex to break up RNA From textbook pg. 290 Back to Chapter 8
Chapter 9: Genome Evolution DNA Covid-19 with solid fill Withering Tree with solid fill Genetic Variation Genetic Variation Life s Family Tree Life s Family Tree Viruses and MGEs Viruses and MGEs Back to Table of Contents
Genetic Variation - Basic Types of Mutations: Within a gene, within a regulatory region, gene duplication, Exon shuffling, Transposition of MGEs, Horizon gene transfer. - In sexually reproducing organisms, only changes in germ line/ gametes are passed on to the offspring. - Point mutations are changes in the nucleotides: add, delete, swap - Mutation in genes are due to natural selection and evolutionary pressure - An example of regulatory region mutation ins lactose tolerance. Lactase production is a mutation of the repressor regulator in the Lac gene. - Gene duplication and divergence can give rise to gene families. An example of this is the duplication of - and - globin genes, each of these then being duplicating and diverging to form hemoglobin protein. This phenomena can lead to whole genome duplications as seen in several plants. - Exon shuffling: Exons from different genomes converge with one another to form family proteins. It is estimated that nearly all human proteins come from these shuffling and duplication methods. - Transpositions of MGEs: DNA sequences that move from one chromosome to another and insert themselves into a genome to mutate the gene. - Horizontal gene transfer: Acquiring genes from different species. Can happen in bacteria and cause antibiotic resistance. Textbook pg. 299
Lifes Family Tree From textbook pg. 314 - Homologous genes: Similar in nucleotide sequence due to their common ancestry. - Genetic changes that provide an advantage are likely to be preserved- natural selection - Phylogenic Tree: A diagram that showcases evolutionary relationships between different organisms. The lines diverge at last common ancestor between two organisms. - 50% human genome is Mobile Genetic Elements and only 2% is coding genes. This gives rise to many conserved areas in across all species such as rRNA - Genome comparisons show that in vertebrates lose and gain DNA rapidly - Exons are similar across conserved areas, introns differ. - Individuality in people is due to changes in SNPs- single nucleotide polymorphisms.
Viruses and MGEs - Viruses change genes in the body by inserting mobile genetic elements. - MGEs are also called transposons. Transposase mediates the movement of transposons. Retrotransposons use reverse transcriptase to convert RNA back into DNA and insert themselves into genomes. They make more of themselves by using this process. Transposons can either remove exons or insert their own MGEs. - Viruses are either DNA/RNA and single stranded or double stranded. They require a protein coating to move their genetic code across cells and infect them. They use host cells and their mechanisms/ proteins to replicate. - Retroviruses are reverse transcribed like Retrotransposons. They are reverse transcribed into DNA and are copied into the cell s DNA. This then leads to production of not only the cell proteins, but also viral proteins such as reverse transcriptase which can further continue the process of viral replication in other cells. From textbook pg. 320
Chapter 10: Analysis of Genes Germ with solid fill Test tubes with solid fill Petri Dish with solid fill Rat with solid fill Isolating and Cloning DNA Isolating and Cloning DNA PCR DNA Cloning PCR DNA Cloning DNA sequencing DNA sequencing Gene Function Gene Function Isolating and Cloning DNA Back to Table of Contents
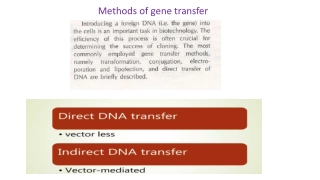
Isolating and Cloning DNA - Restriction enzymes cleave strands of DNA at specific sites and leave sticky ends for different cut DNA to bind to. - Gel electrophoresis separates nucleic acids by size and charge. Northern blot is RNA electrophoresis and southern block is for DNA electrophoresis. Protein electrophoresis is called Western blot. - A vector is something that holds DNA. Plasmids are types of circular DNA vectors that can be ligased and cloned in bacteria. By cleaving genomes with restriction enzymes, inserting them into vectors such as plasmid and transecting it into a bacteria can produce a genomic library. - A cDNA library is collected when mRNA is reverse transcribed into cDNA. This process includes purifying mRNA, hybridizing poly T tails, reverse transcribing RNA and then replicating the single strand of DNA with DNA polymerase. From textbook pg. 340 Back to Chapter 10
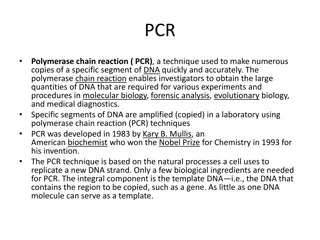
PCR DNA Cloning From textbook pg. 344 - PCR is Polymerase Chain reactions - For PCR, you need double stranded DNA, free nucleotides, primers that can hybridize with the DNA, and a DNA polymerase that can withstand certain temperatures. - Step 1: Heat the two strands of DNA to a particular temperature to separate them. - Step 2: Cool to the right temperature so that the primers are hybridized to the DNA. This hybridization allows the DNA polymerase to start working and replicate the two DNA strands. The polymerase works between the two primer areas. The selectivity of primers allows researchers to clone and replicate particular DNA strands. - Step 3: Continue this process in test tubes for millions of times to clone DNA. - PCR can be used for diagnostic and forensic applications. It can be used to track infections in DNA by amplifying a DNA sample. It can also be used for STRs or DNA fingerprinting, where mRNA is reverse transcribed and amplified by PCR and then run across a gel to compare with control blood samples. Back to Chapter 10
DNA sequencing - Dideoxy sequencing or Sanger sequencing is done by analyzing DNA s terminated chains. - Step 1: Remove the OH group from deoxyribonucleotide. This produces ddNTPs, which are used to terminate DNA replication. - Step 2: Prepare 4 test tubes, with many dNTPs(with OH) and some specific ddNTPs. Each test-tube will have one type of ddNTP such as A,C, T, G. Each test tube will have several dNTPs and only one type of some ddNTPs. - Step 3: By adding DNA polymerase, primers, ddNTPs, and dNTPs, you have random stops of DNA replication. By looking at which ddNTP had smallest to biggest chain, you can sequence the DNA strand. DNA polymerase works 5 -3 so the template strand should be 3 -5 end. - Next generation sequencing is faster, cheaper and more advanced. The process involves labeling ddNTPs which are mixed with PCR amplified DNA and primers. Each time the ddNTP is connected, the florescent dye is activated and is recorded in a machine. Then the tag and terminator ddNTP is removed and the process continues. From textbook pg. 347 - You can use the sequences of DNA of two different proteins to determine their shape, function and family type. Back to Chapter 10
Gene Function - In situ hybridization uses complementary or antiparallel DNA strands with labeled florescent dyes to locate where a specific RNA/DNA sequence is. The sequence of a florescent protein(GFP) can be added to a sequence to light it up and make it visualized. - Classical Genetics: Mutating many different genes by chemicals or drugs and analyzing the effect on the role of a protein. They tell us if a protein is necessary and if the cell can work without it. - Reverse Genetics: Targeting specific genes by using RNAi to destroy complementary mRNA and reducing protein production. The goal is to see if the protein is necessary for cell s survival. - Regulatory or expressed genes can be modified by transfecting embryonic cells with known genes and implanting them into an organism. - CRISPR: A combination of Cas9 protein and guide RNA that can target specific gene and alter or terminate it. Used for gene expression as well. - Transgenic plants can be beneficial for humans. Not all GMO plants are transgenic. From textbook pg. 358 - Rare proteins such as hormones can be made to replace missing hormones by cloning recombinant DNA. Vaccine antigen can be created by cloning specific DNA sequences. Back to Chapter 10
Work Cited Alberts, Bruce, et al. Essential Cell Biology. 5th ed., Cengage Learning, 2018.