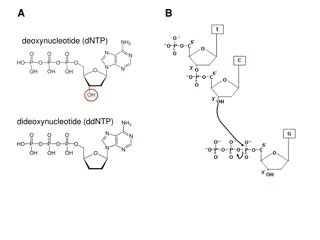
Pulsed-Field Gel Electrophoresis: Separating Large DNA Molecules
Pulsed-Field Gel Electrophoresis (PFGE) is a technique developed to effectively separate large DNA molecules through the application of an electric field that periodically changes direction. This method, introduced by David C. Schwartz and Charles C. Cantor in 1984, revolutionized the resolution of DNA fragments larger than 15-20kb. PFGE uses restriction enzymes to cut bacterial DNA at specific sites, generating fragments that can be separated based on size. The process involves loading treated DNA plugs onto an agarose gel and applying an electric field to separate the restriction fragments. Unlike traditional DNA separation methods, PFGE can effectively separate multiple large DNA fragments, allowing scientists to create DNA fingerprints for bacterial isolates.
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author. Download presentation by click this link. If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
E N D
Presentation Transcript
December 17 pankajgaonkar12993@gmail.com 1
Historical background of PGFE Standard gel electrophoresis techniques was unable to separate very large molecules of DNAeffectively. DNAmolecules larger than 15-20kb migrating through a gel will essentially move together in a size-independent manner. At Columbia University in 1984, David C. Schwartz and Charles Cantor developed a variation on the standard protocol by introducing an alternating voltage gradient to improve the resolution of larger molecules. This technique became known as pulsed-field gel electrophoresis (PFGE). 2
WHAT IS Pulsed Field Gel electrophoresis ??? Pulsed field gel electrophoresis is a technique used for the separation of large deoxyribonucleic acid (DNA) molecules by applying to a gel matrix an electric field that periodically changes direction. PFGE is a technique used by scientists to generate a DNA fingerprint for a bacterial isolate. Pulsed Field - any electrophoresis process that uses more than one electric field alternating. Fig.-A microbiologist runs a pulsed-field gel electrophoresis test used in bacte3 rial typing.
How does PFGE work? PFGE uses molecular scissors, called restriction enzymes, to cut bacterial DNAat certain locations known as restriction sites. These molecular scissors are selected to generate a small number of DNApieces that can be separated based on size. Usually these DNA pieces, or restriction fragments, are large and need to be specially treated and separated to generate a DNAfingerprint. First the bacteria are loaded into an agarose suspension, similar to gelatin, then the bacterial cell is opened to release the DNA. Once the DNAis released then the agarose and DNA 4
Continued The treated plugs are then loaded onto an agarose gel and the restriction fragments are separated based on size using an electric field. What makes PFGE different from how scientists usually separate DNAis because PFGE can separate several large restriction fragments. T o do this an electric field that constantly changes direction to the gel is used to generate a DNAfingerprint. 5
The Pulsed-feld Ge i l Electrophores is Process Pulsed-field Gel E l ectrophoresis (PFGE) Cut DNA with Restriction Enzyme e The bacterial cells are broken open with blochemlcaJs, or lysed, so that the DNA is free in the agarose plugs. The scientist takes bacterial cells from an agarplate. 0 The scientist loads the DNA gelatin plug into agel. and places i11nan electric field that separates DNA fragments according to theirsize. DODD Plug Mold Data Analysi s (BioNumerics)e The gel isstained sothat DNA can beseen under ultraviolet (UV) light Adigital camera takes a photograph of the g-el and stores the picture in the computer. e The scientist mixes bacterial cells with melted agarose and pours into a plug mold.
APPLICATION PFGE may be used for genotyping or genetic fingerprinting. It is commonly considered a gold standard in epidemiological studies of pathogenic organisms. Subtyping has made it easier to discriminate among strains of Listeria monocytogenes and thus to link environmental or food isolates with clinical infections. 7
Advantages of PFGE PFGE subtyping has been successfully applied to the subtyping of many pathogenic bacteria and has high concordance with epidemiological relatedness. PFGE has been repeatedly shown to be more discriminating than methods such as ribotyping or multi- locus sequence typing for many bacteria. PFGE in the same basic format can be applied as a universal generic method for subtyping of bacteria. (Only the choice of the restriction enzyme and conditions for electrophoresis need to be optimized for each species.) DNA restriction patterns generated by PFGE are stable and reproducible. 8
Limitations of PFGE Time consuming. Requires a trained and skilled technician. Does not discriminate between all unrelated isolates. Pattern results vary slightly between technicians. Can t optimize separation in every part of the gel at the same time. Don t really know if bands of same size are same pieces of DNA. Bands are not independent. Change in one restriction site can mean more than one band change. Relatedness should be used as a guide, not true phylogenetic measure. Some strains cannot be typed by PFGE. 9
Video Link: https://www.youtube.com/watch?v=__QhCX12h8I Reference Book: Wilson and Walker's Principles and Techniques of Biochemistry and Molecular Biology
December 17 pankajgaonkar12993@gmail.com 10