Understanding the Morphology and Genome Organization of NDV
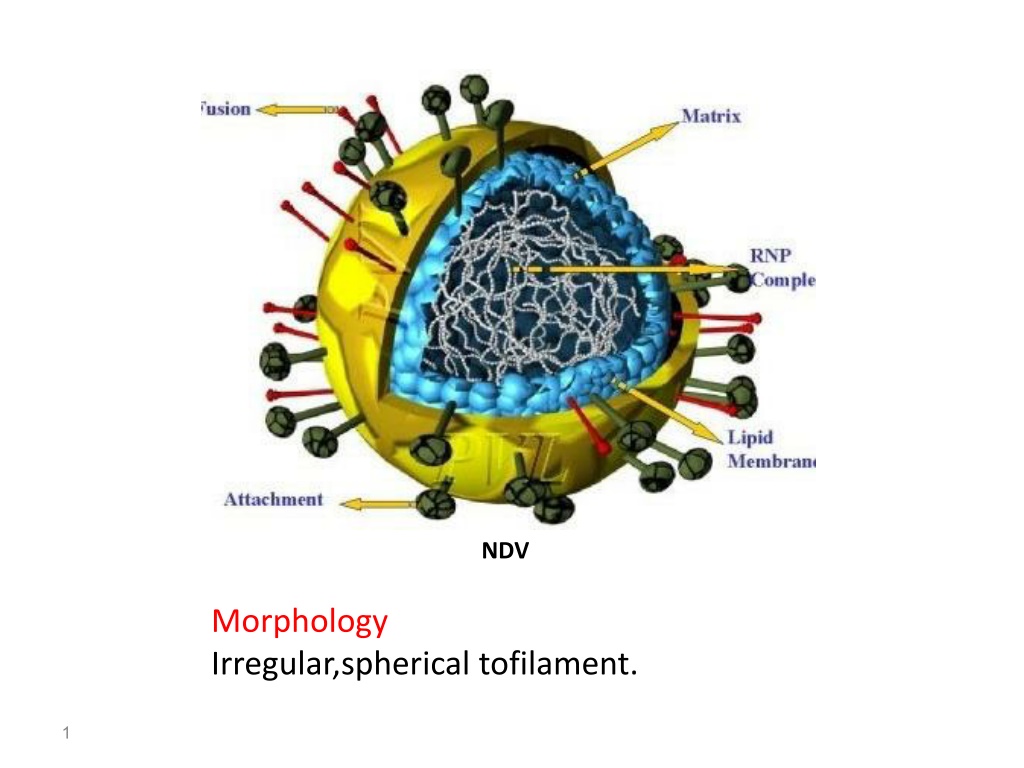
NDV, a pleomorphic virus, has an irregular spherical shape with filamentous features. Its genome consists of a single-stranded RNA with six genes arranged in a specific order. The virus contains a lipoprotein envelope with short spikes and a helical nucleocapsid core to protect the RNA. The genomic organization includes open reading frames encoding various proteins, with additional non-structural proteins generated by RNA editing. Various sequences within the genome play crucial roles in transcription and replication. The classification of NDV places it under the genus Avulavirus in the Paramyxoviridae family due to its specific affinity for mucopolysaccharides.
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author. Download presentation by click this link. If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
E N D
Presentation Transcript
NDV Morphology Irregular,spherical tofilament. 1
Size,150 to300 nm in dimeter so it is pleomorphic. 1.the lipoprotein envelope covered with short spikes(Epitopes) glycoprotein in nature. 2.Matrix protein at inner side of envelope; is paracrystalline layer to stabilize virus structure. 3.Helical symmetry Nucleocapsid core (18)nm in diameter.it protect RNA aganist ribonuclease attack. The Helical nucleocapsid core is composed of the genomic RNA, nucleocapsidproteins(N), phosphoproteins and polymeraseproteins. 4.RNA Is (ss RNA) ,NON segmented,polycistronic-ve sense, 15,2kb in length. 5.RNA genome is 15,186 nt long and consists of six genes in the order of 3 leader sequence N-P-M-F-HN(attachment)-L 5Trailer sequence. 6.The genome contain (55)nt leader sequence at (3)end act as transcriptional promotor, and(144)nt trailer sequence at (5)end. 7.Leader sequence and Trailer sequence are (Extracistronic NON CODING Regions and not copied into mRNA. 8.There are intergenic sequences are (2,1,1,31,47) nt between N\P ,P\M,M\F,F\HN,HN\L genes respectively(Determine the genus) 4
Genomic Oraganization(acc to Alexander D.J NDV RNA genome consists of 15,186 or15,192 or15,198 (nt). It contain six open reading frames(ORFs) which encode NP,P,M,F,HN,L. Two additional non structural proteins (V,W) which generated by RNA editing during (P) gene transcription pprotein encoded by an unedited transcript of the P gene, whereas the V and W proteins are the result of an mRNA-editing event in which one (V) or two (W) G residues are inserted at a specific position within the P gene mRNA.(Jornal of General Virology(2004),85,2375-2378 5
Exact complementarity found between the first &last eight nt of genome (identical in all NDv strains ) this indicate that this regions important in transcription&replication. Conserved transcriptional control sequences of 10 or 11nt are preceding &follow each gene. 6
Gene start sequences act as transcriptional promotor. Gene end seq. Transcriptional terminator. Gene Gene-start Gene-end NP ACGGGTAGAA TTAGAAAAAA P ACGGGTAGAA TTAGAAAAAA M ACGGGTAGAA TTAGAAAAAA F ACGGGTAGAA TTAGAAAAAA HN ACGGGTAGAA TTAGAAAAAA L ACGGGTAGAA TTAGAAAAAA 7
Classification of NDV acco to international committee on Taxonomy. Para(beyond), myxo(mucous)in Greek.mean that specific affinity of paramyxoviridae for mucopolysaccarides and glycoproteins particulary sialic acid containing receptors on the cell receptors of Upper respiratory tract. Virus by Latin (poison). Group;V (-ssRNA) Order;Mononegavirales. Subfamily;Paramyxovirinae. Genus;Avulavirus. Species;Avian Paramyxovirus serotype;Avian paramyxovirus serotype 1 8
Serotypes of avian paramyxovirus. 1.Avian Paramyxovirus-Type 1(APMV-1).(NDV) 2.APMV-2 3.APMV-3 4.APMV-4 5.APMV-5 6.APMV-6 7.APMV-7 8.APMV-8 9.APMV-9 10.APMV-10 11.APMV-11 9
APMV-1has gene map structure similar to rubulaviruses,which the reason for initial classification. Further analysis discovered that unlike other Rubulaviruses. It lacks (c) protein ,small hydrophobic protein(sH) . Phosphoprotein(p)relatively small. The intergenic region is also variable compared to other rubulaviruses The nucleotide sequence not align with rubulaviruses nor respiroviruses,so APMV-1 classified under new genus(Avulavirus). 10
Viral proteins; The genome encode six structural proteins(NP,P,M,F,HN,L) 11
1.The core proteins;(complexed with the virion RNA). (Helical nucleocapsid core) consists of 1-N gene encode NP protein that bind to genomic and antigenomic RNA. 2-P,L genes encode P,L proteins bind to NP to form RNP complex.P is cofactor for L protein(mediate binding ofL with nucleocapsid. 3-L;catalytic subunite of RNA dependant RNA polymerase; it responsible for Replication,Transcription,Regulation of R.C 4-M gene encode M Protein required for virus assembly &rigidity. 2.Surface glycoproteins;(associated with the lipid enveloped) 1.F gene encode F glycoprotein .(fusion,pathogenesis,virulence,neutralizingAgs) 2.HN gene encode HN glycoprotein(Attachment protein). They are required for viral attachment, pathogenesis,virulence,and also neutralizing &protective Ags(Recombinant vaccine) 12
Replication cycle of NDV. 1-Attachment. 2. Penetration by fusion. 1. First step of Replication is attachment of virus with host cell membrane ,HN protein bind to (SA) receptors on the surface of the cell membrane bring (F)protein close to cell membrane. 2.HN interaction with (SA) receptors intiate conformational changes to activate (F)protein. 3. during fusion events F1 polypeptide undergo addional conformational changes which expose RHA,RHB regions. 4. the two hydrophobic regions act to bind viral membrane to host membrane. 5. the N terminal fusion peptide attach to host cell membrane,the transmembrane domain anchors the viral membrane. 6.a 6 helix bundle couples free energy released during protein refolding when two membrane merge. 7-the final conformational state of F protein is the most stable form and not reversible. 13
8.The membrane fusion occur at neutral PH,but exact mechanism of fusion activation unknown. 9.the accepted steps during fusion events begin with docking of viral membrane to host cell membrane due to interaction between HN,SA. 10.F protein is activated as the membranes approach and upon membrane merging pore between membranes. 11. F protein not sufficient alone for virus fusion,it need HN attachment to promote fusion. 12.NDV isolates have mutations in attachment function of HN to continue to promote fusion 14
3- Uncoating;RNP complex enter host cell. 4-Replication of nucleic acid ;in cytoplasm as the genome is ve sense RNA so L required to enter host cell with its RNA(Viral genome encodes its RNA dependant RNA Polymerase)which act as Transcriptase &Replicase. A-Transcription to mRNAthat translated into viral protiens (non structual&structural). B-by replicase enzyme(L) ;form complement +SSRNA act as template for production of newRNA viral progny. 15
Transcription of RNA . 1.RNP complex is template for transcription by RNA dependent RNA polymerase L protein. 2.L protein binds genomic RNA AT (3)entry site of RNP complex and transcribes the six protein genes using start-stop mechanism. 3. there are gradient of Mrna synthesis inversely related to distance from 3 end of genome. 4.protein gene proximity to 3 end result in high production of protein. 16
After primary transcription &translationwhen sufficient amounts of unassembled N protein are present, viral RNA synthesis becomes coupled to the concomitant encapsidation of the nascent [+] RNA chain. Under these conditions, vRNAP ignores all the junctions (and editing sites) to produce an exact complementary antigenome chain in a fully assembled nucleocapsid 17
Translation of mRNAto protein; 1. Fusion glycoprotein. Is class (I) Fusion glycoprotein is synthetized as type(I) integral membrane protein. When the protein translated three identical polypeptide chains assemble into homotrimers then CHO chains is added (inactive),then by host protease cleave the precursor protein to be active. 18
-F protein cleavage site differ according to virulence of NDV strain. -Synthesis of F protein occur along ER as inactive F Precursor so it must be activated by cleavage to F2,F1 polypeptides to be infective.they must remain bound to the viral surface by disulfide bonds to be infectious. -F1 polypeptide has two hydrophobic regions are(N terminal fusion peptide&transmembrane domaine). -It also contain two heptad hydrophobic repeat regions HRA&HRB. -upon intial translation F protein folds into metastable form prior to fusion,after activation occur large scale conformational changes,these changes progress down energy gradient to form stable postfusion conformation. -active F1 polypeptide mediate fusion between viral lipid membrane&host cell membrane,so entry of viral genome,so replication. 19
2.HN surface glycoprotein;is type (II) integral membrane protein.play important role in; 1.attachment. 2.entry. 3.Tropism. 4.virulence (Reverse genetics procedures). 5.neutralizing&protective Ag.(against it produced neutralizing&protectiveAb. 20
5-Assembly;appropriate conc.of virus particles and nucleic acids transported and localized at plasma membrane where occur self or spontanous assembly of protomers to form capsomers in helical symetery to form capsid that package NA (nucleocapsid) . - host cell membrane modified to form viral envelope. -during viral assembly ,viral surface glycoproteins expressed on the surface of host cell membrane so intiate fusion between neighboring cells so cell to cell transfere so cell syncytia with F protein without aid of HN protein . This step under direction of M protein. 6-Maturation;in this stage the virus become infectious -the cellular protease cleave F protein to be infectious and active. 7- Release; by cell to cell transfere to form cell syncytia&by budding through cell membrane and make cell damage. 21
Classification of NDV. Two different classification based on genetic analysis. First classification; proposed by aldous et al based on genotypes or genetic lineage grouped under serotype 1. Second classification; based on genomic characterization& sequence analysis of F,L genes. Other classification; 1. according to antigenicity; one serotype.(Abs produced against one strain(pathotype)can protect against other strains. 2. according to virulence(pathogencity); 1.Virulent strains. -Velogenic(severe virulent)strain. -Mesogenic(moderate virulent)strain. -Lentogenic(mild virulent)strain. 2.Avirulent(asymptomatic)strain. 22
Virulence determinants; 1. Genetic composition of F protein cleavage site.(see classificaton&DNA sequence. 2.AAs differences in HN protein.(Reverse Genetics Procedures). 23
Vaccines; 1.Live attenuated; 1.Lentogenic strains; HitchnerB1 ,F,Lasota.(ACCto ICPI) By serial passages on ECE by allantoic route till lost its virulence to original host. They are alive and able to infect cell ,it mimic the natural infection and induce all three imune responses(cell medited immunity,humeral immunity,secreted Ab producing mucosal immunity. -Age;young&also in all ages, -Dose;10^6.5 and 10^7 EID50 per bird. -Route; eye or nostrile drop or aerosol or drinking water. -Lasota cause post vaccinal respiratory signs ,used as booster vaccine in flocks vaccinated with F,B1. 24
2.Mesogenic strains;Komarov,mukteswar,Rokain(accor to ICPI) Age ;not less than 8 week,not in partially immune birds and not in birds not previously immunized to avoid death. -Dose;10^5 EID50. -ROUTE ;injection. -used as booster vaccine. 3. Avirulent strains;V4,V4-HR,1-2 -All ages. 2.Killed attenuated vaccine. Velogenic strains; BeaudetteC,GB Texas,Herts33.(ACCO to ICPI) attenuated as live vaccineThen killed by beta propionolactone or formaline(inactivation of protein) then add Adjuvant(aluminum hydroxide but now use oil emulsion (as it stimulate immune system rapid than Alluminum hydroxide). 25
3.Virus vector vaccine; by using Fowl pox or Marek s disease virus as avector. -remove F,HN genes -remove virulent genes of Marek s virus provided that not affect on replication and insert F,HN genes then injected in bird as avaccine. It is marker vaccine used to differentiate naturaly infected bird from vaccinated 26
Diagnosis; 1.provisional diagnosis;relatively non specific signs and PM lesions so. 2.confirmatory laboratory diagnosis; 1.sampling . 1.collecion. 2.preservation. 3.preparation. 2. isolation. SPF ECE, Allantoic sac route. 3.identification. 1. non serological tests; -HA,infectivity. 2.serological; HI, serum neutralization,AGPT,ELISA. 27
Samples; (acc to Developments in vet virology) Newcastle disease. Author;Dennis.J.Alexander. For succesful isolation,res&intestinal tract. 1.live birds show clinical signs 1.faecal sample. 2.cloacal swabs 3. tracheal swabs. 2. Dead birds; acc to organs obviously affected Lung liver spleen kidney brain. 28
Most NDV isolates stable in non putrifying tissue & organs samples or faeces providing that not expose to high tep. In low tep countries ,no problems in sample transportation,but in tropical countries occur deterioration of sample so Bone Marrow is useful sample for virulent strains as can dable to demonstrate virus after several days at 30c 29
transport Media, -con of antibiotic vary from lab to lab due to local conditions &availability, not appear to be critical . Typical sample. 10,000U\ml penicillin,10mg\ml streptomycin,250microg\mlgentamycin 5000u\ml mycostatin.in addition 50mg\mloxytetracycline if presence of chlamydia 30
Preparation; 1.Tissues&Organ samples finely minced and 10-20% w\v suspensions of them and faeces samples made in phosphate buffer saline contain antibiotics,PH 7-7.4. 2. swabs placed in sufficient antibiotic PBS to allow full immersion. 31
The samples should held at room tep for about 2hr before centrifugation at 1000g. Virus isolation, most virulent strains of NDV grow in wide range of cell culture due to local conditions. It is the best method ,but the most widely recommended method is treatment of sample with antibiotics & inoculation in ECE SPF 9-10 days old ,or at least eggs from hens free of NDV antibodies. -Steps of isolation -harvestation .. -detect signs on embryo,fluids,membranes(multisigns system). -Detection by HA Test on fluid(after purification and concentration by centrifugation then collect supernatant and discard sedement. 32
Viral Detection; 1.HA test; on harvested allantoic fluid by using susptible con washed Packed RBCs (wide range). 2.infectivity test todetermine EID50. 3.IFT; Immunofluorescence staining of tracheal section or smear. Viral identification; serological tests; 1.HI test,on heamagglutinag harvested allantoic fluid by using referance known sera,it is the test of choice due to rapid result. 2. SNT. 3.IFT.some what less sensetive. 4. ELISA. Also serological tests used for surveillance of NDV, -monitor vaccinal immunity, -vaccine potency. 33
Molecular identification of different genes. 1.Rrt-PCR to determine the virulence and strains. 34
Virus characterisation; ICPI; IVPI; MDT. 35