Understanding Transcription: The Initial Steps in Gene Expression
Gene expression involves two crucial phases - transcription and translation. Transcription, the first step, is the process where RNA is synthesized from DNA with the help of RNA polymerase. It begins with initiation, where the enzyme recognizes the promoter region and forms a complex with the DNA, moving on to elongation where RNA synthesis occurs along the template. This process is essential for gene regulation and protein synthesis.
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author. Download presentation by click this link. If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
E N D
Presentation Transcript
TRANSCRIPTION By S. Sivaranjani Arunnehru Assistant Professor Bon secours college for women Thanjavur
INTRODUCTION Presence of gene in the genome is not sufficient to express as proteins. Genes proteins Gene Expression It is divided into 2 phases Transcription or RNA synthesis Translation or protein synthesis Transcription: It is the first step in gene expression and is predominant in level of gene regulation It is another type of Nucleic acid produces another type of it in the process of RNA synthesis from DNA molecule with the help of complex protein Concept: mRNA synthesis Carry information form gene located in nucleus to ribosome to cytoplasm (Crick, 1958).
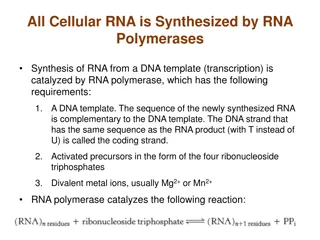
BASIC MECHANISM OF TRANSCRIPTION: INITIATION: RNA polymerase enzyme recognize promotor region lie just upstream of gene and bind to it. Initial structure formed between RNA polymerase and DNA closed promoter complex. Enzyme covers or protect about 6 bp of double helix starts from just upstream of 35 box and down stream of 10 box. 10 box the DNA helix unwinds due to breakage of bp [melting]. Melting is an essential prerequisite for transcription : bases of template strand must be exposed in order to discrete transcription. Synthesis of open promoter complex is forced I ST 2 ribonucleotide are base paired to template polynucleotide at position +1 and +2 and I stphosphodiester bond of the RNA molecule is synthesized by RNA polymerase. Transcription follows same base pairing rules as DNA replication.
T, G, C, U and A in DNA pair. Bp pattern ensures that the RNA transcription is faithful copy of gene. End of initiation signified by promoter. Clearance, where RNA polymerase moves away from the promoter site without dissociating, freeing the promoter from further initiation events. Promoter clearance occur only if the open promoter complex is stable and follows a number of abortive initiation where small transcription are generated. Initiation rate limiting steps in transcription and high level of gene regulation in both prokaryote and eukaryote.
ELONGATION: RNA polymerase moves along the template synthesize nascent RNA in 5-3 direction. The DNA is unwound ahead of elongation complex and rewound behind dynamic , melting structure represent the site of transcription trans bubble. Transcription bubble between 12 to 17 bp in length and is constant through out transcription. RNA polymerase unwindase and windase activity as no other enzymes such as helicase or topoisomerase taking part in transcription. Polymerization- 3 oh group pf one ribonucleotide reacts with 5 po4of 2 nd ribonucleotide to form a phosphodiester bond- loss of pi for each bond formed. The chemical reaction modulated by the presence of DNA template directs the order in individual ribonucleotide are polymerized into RNA.
As polymerase move along , it must insert the proper nucleotide into growing chains at each site.the enzyme is capable of selecting the complementary ribonucleotide triphosphate for incorporation as a result of ability of nucleotide to form the proper stereo chemical fit with nucleotide in DNA strand being transcribed . Bacterial RNA polymerase is capable of incorporate 50 nucleotide/second into growing RNA. New RNA transcribed from 5 to 3 direction . New ribonucleotide being added to free 3 end of existing polymer. The length varies from gene to gene or it may be short as 20 nucleotide or longer than 600 nucleotides .
TERMINATION: It is not a random process, but must occur only at a suitable position shortly after the end of all gene. Polymerase moves down to the DNA until RNA polymerase reached a stop signal or terminal sequence. Termination occur with the help of rho protein is known as rho depended termination and which occur without the help of rho protein is called rho- in depended termination. Both rho depended and rho in depended termination involve the formation of stem loop. RNA polymerase pauses at the stem loop structure, rho catches up to the polymerase and unwind the DNA RNA hybrid . rho can do it has DNA RNA helicase property. Yet , exact segment of events at the terminator is not fully known presently.
TRANSCRIPTION IN EUKARYOTES: It is a complex process- one group of RNA polymerase and associated proteins are involved in this process. Different RNA polymerase exist for different types of gene. Process of eukaryotic transcription is less understood because certain RNAs and mRNAs have different lifespans in cell complicate the situation and mammalian cell different from prokaryotes.
RNA POLYMERASE: There are 3 different types of RNA polymerase: RNA polymerase I: nucleolus transcription of genes for rRNA. RNA polymerase II: synthesis of heterogenous nuclear RNA or mRNA. RNA polymerase III: transcription of small nuclear RNA and t RNA. Size of RNA polymerase- Molecular weight 500,000 to 600,000. All eukaryotic RNA polymerase have similar structure and to extent have same AA segment but respond to one peptide toxin [ amanitin] in different ways. amanitin blocks the translocation of RNA polymerase during transcription . RNA polymerase I insensitive to amanitin but RNA polymerase II is sensitive even to lower concentration, where as RNA polymerase III is sensitive at high concentration of - amanitin.
Eukaryotic polymerase II consists of 14 subunits . Carbonyl terminal repeats domain [CTD] -26 repeated units in yeast and 52 units in mammalian cells . Promoters: Region in which RNA polymerase binds and begin transcription. In eukaryotic , promoters do not start frequently events to occur. 3 different RNA polymerase in eukaryotic nuclei. More than 3 different promoters recognized by different polymerase. RNA polymerase does not recognize eukaryotic promoters by itself. For the recognition RNA polymerase require help or relies on proteins and transcription factors.
Promoters of RNA polymerase II: polymerase II responsible for transcribe the vast majority of gene coding for proteins. Confer fidelity and frequency of initiation and rigid require for both position and concentration . single base changes have dramatic effect on functions. RNA polymerase III: Promoters that governs 5S transcription of 5s rRNA and tRNA genes transcription different from bacterial polymerase. III promoters located within the genes they enter. Promoters of +50 to +83 bp from start point of transcription it is a internal control region.5s RNA gene promoter split into A and C box with short segment in middle- intermediate element. tRNA , ICR split into A and C box without any intermediate element sequence between 2 boxes is of least important transcribe gene encoding other small RNAs contain promoters and within gene but at 5 end of gene starting. One small RNA TSL RNA contain 1 TATA box.
RNA polymerase I: base sequence not match with TATA or CAT box. It recognize promoters present on rRNA gene. Promoters of this gene present at the 5 end of gene. Ist promoters core element between -45 and +20. This promoter segment is conserved like TATA box and 65 bp in length. Other found between -156 to -107 before transcription site upstream control element [UCE] 49 bp in length.
TRANSCRIPTION FACTOR: Protein assist RNA polymerase in recognize promoters. Two groups: Activate specific genes: gene specific transcription factors or gene activation. Activate any gene: general transcription [GT] factor GT bind DNA region within promoters by themselves and deliver RNA polymerase to promoter site. GT occur at low level or basal level but gene specific transcriptase can act both +ve[boost the transcription rate] or ve [inhibit transcription rate].
RNA polymerase I transcription factor: Pre initiation complex forms at rRNA promoters involves binding if polymerase I in addition to 2 transcription factors called SL-4 and upstream binding factor [UBF] to the promoter. UBF binds to upstream control element of rRNA promoters and causes polymerase I to bind near the transcription start site. UBF binds SLI join complex strength polymerase I binding promoter- formation of pre initiation complex. SLI complex protein with a peptide- TBP one of protein in TATA binding protein family. Similarly to TFIID and 3 peptides bind to TBP 3 peptide TAF3 and similarly to TAF5 in TFIID.
RNA polymerase III: transcribe different type of genes , transcription complex are also different transcription complex are same but complex formed are different. 3 factors involved in 5s rRNA transcription: TFIIA,B AND C TFIIB true transcription factor others are assembly factors. It helps RNA polymerase III binds just down stream in a position to start transcribe the gene. TFIIIB and C bind to internal promoter followed by TFIIIB. TFIIIB position RNA polymerase III at transcription start site. tRNA gene transcription 2 factors- TFIIIB and C binds internal promoter. TFIIIC protein protein interaction. TFIIIB contains TBP like S1 and same role- SL1. TFIIA and C Same role as UBF.
RNA polymerase trans factors: it is a complex interact with many factors before they initiate the transcription. 3 classes of transcription factors- regulation of transcription by RNA polymerase II. Istclass basal factors . Ex: TBP, TFIIA etc.,- basal transcription .it has number of factors-TFIIA, B,D, E, F, H. TFIID- complex protein consists of TBP and TAFS 700 KD protein covers a 35 bp region of DNA. TBP 20-40 KD protein binds to 10 bp segment of DNA over TATA box basal transcription. TFIID- support enhanced and basal transcription. 2ndclass co activators .ex: TAFs enhanced transcription co activators.
Different combination of TAFs with TBP bind to different promoters and activate them with different strengths. TAFs- basal promoters- initiation complex interacts upstream activators- basal promoters-cell type specificity, enhance transcription rate convert basal level transcription enhance the level of activators. Sequence element increase or decrease thee rate of transcription. initiation are enhancer or repressors. Enhancers supress the transcription-silencers or repressors. Enhancers or silencers are same in structure except function. enhancers are DNA sequence present hundreds of bp away from start of gene and enhance transcription stimulate transcription from promoters.
INITIATION: Formation of basal transcription complex. Recognition of TATA box by TFIID factor. TBP makes sequence- specific contacts with DNA at TATA box through minor groove. Most transcription factors bind to major groove. Role of TAFs form an assembly that directs the formed pre-initiation complex to promoters. 1. TFIIA- Istblocks binding of DRI inhibits TFIID activity. TFIIB binds and act as a bridge protein for TFIIF carries RNA polymerase II into complex. TFIIB and F together promote specific interaction between RNA polymerase II and the start site. Enzyme facilitate binding of TFIIE allow bind of TFIIH and J. H- important pre initiation complex due to helicase activity controls promoters melting and kinase activity. phoslates the CTD of RNA polymerase. Addition of TFIIE and M final assembly of pre initiation complex or basal transcription complex.
Binding event extends the size of complex, so 60 bp. Binding of TFIIH- release the enzyme from the IC and facilitates promoter clearance leaving IC at the promoter like bacterial RNA polymerase. Eukaryotic RNA polymerase abortive initiation before promoter clearance. Istnucleotide- purine modified by mRNA guanyl transferase to generate a cap.
ELONGATION: Encounter secondary structures lead to arrest or termination of transcription . To prevent, various elongation factors are associate with RNA polymerase during elongation site side by side. Elongation is slower when compared to bacteria. Transcription is longer in the presence of long gene. Ex: human dystrophin gene- largest gene- transcription of 2.5 mega base pairs takes 16 hours to complete.
TERMINATION: Poorly characterised. RNA polymerase II transcription are processed by polyadenyl at 3 OH end. Termination is not clear. Mechanism of transcription by RNA polymerase by RNA polymerase I and III: Transcription of rRNA genes by RNA polymerase I and 5S rRNA , tRNA genes by RNA polymerase II is similarly to that of RNA polymerase II with minor difference in transcription factor initiate pre initiation complex formation . Elongation is same. But termination is different from prokaryotic termination and eukaryotic RNA polymerase II transcription termination. Termination of RNA polymerase II transcription occurs at a site 15 bp to the end of matured RNA and involves recognition of a specific cis- acting element. Termination of RNA polymerase III transcription occurs at sites similarly to bacterial rho- independent terminators.
POST- TRANSCRIPTIONAL MODIFICATION: Transcription and translation not occur simultaneously, occur at 2 different sites. Eukaryotic transcription result in primary transcription. 3 major changes occur in the primary transcription before it is transport into cytoplasm for translation. 1. cleavage of introns. 2. addition of t-methyl guanosine gps or mRNA capping. 3. poly[A] tail.
MRNA CAPPING 1stprocess. Cap- 6 ethyl guanosine attach backward through tri phosphate linkage to 5 terminal end of mRNA . The nuclear enzyme guanyl transferase or mRNA guanyl transferase catalyses addition of guanosine triphosphate part of cap. This type of cap is called 0 capping. Associate with RNA polymerase IC guanine 7- methyl transferase present in cytosol methylates this guanine. Methyl group comes from s- adenosyl methionine and methyl group at 7 position of guanosine. In higher eukaryotes methyl group transfer the position of o2of ribose sugar in next nucleotide. i.efirst nucleotide in transcription corresponds to position to generate a type 1 cap ex: human.
2 methyl groups present one on the guanosine and other is sugar of the other next or +1 residue. +1 after guanosine is adenine methyl group N6position of adenine o2of sugar. Type 2 cap. Triphosphate does not link to 3 with 5 .instead the 2 end nucleotide link through 5 sites. Capping of nucleotide back in to join the growing RNA chain. Capping occur before transcription so it is called as co transcriptional process. 2 functions: 1. it protect mRNA degrade by RNAse 2. important to bind mRNA to ribosomes
POLYADENYLATION: Most eukaryotic mRNA chain of 40-200 adenine nucleotides attach to 3 end .poly [A] tail is at transcribed from DNA rather add after transcription by nuclear enzyme poly [A] polymerase. Tails stabilize mRNA and facilitate their exit from nucleus after mRNA enters cytosol, poly [A] tail is shortened. Multi subunit complex trimeric cleavage factors cleavage and polyadenylation carry out cleavage reactions. enzyme polyadenylation polymerase[PAP] catalyses the addition of adenylate residue. Polyadenylate binding protein attaches to tail and increases the processivity of PAP. Controls the maximum length of polyadenylate tail. Cleavage of polyadenylation complex: 2 enzymatic activities: endonuclease and poly[A] polymerase. Poly [A] site- with the signal AAUAAA and downstream GU/U region . A complex of CPSF , CFI and II and CS+F bind to these sequence. Cleavage occur and CPSF remains and joined by PAP- synthesis of poly[A] result in addition of 1STIOAs.
PAB II join the reaction stimulate the synthesis of poly[A] extend the tail-200 A residue 2 functions of poly[A] mRNA protection and translatability. Function of poly[ A] : Cap increases the translation 300 fold poly[a] increases the translatability of message by about 20 fold. RNA splicing: Removal of introns [non coding elements present in RNA molecule] Higher eukaryotes- non coding introns separate coding sequences or exons. Primary transcription: entire sequence of gene and non coding sequence are spliced out during processing.
Splicing mechanism join exon sequence to signal nucleotide codons in exon distal to introns- correct reading frames. 3 introns excision from RNA transcripts. Splicing of tRNA precursors: excision of introns from yeast tRNA precursors in 2 stage. 1st- nm bound endonuclease makes 2 cuts at end of introns. Spicing ligase joint the 2 halves of tRNA to produce neutral form of tRNA molecule. Intron removed- multiple steps. 2nd- covalent modification of substrate RNA is cleaved at precise 5 site- ligation.
AUTOCATALYTIC SPLICING OF TETRAHYMENA: Autocatalytic excision of intron in the tetrahymena are RNA precursors no external energy and no proteins Series of phosphodiester bond transfer no bond lost or gained in process. The reaction require a guanine nucleotide or nucleoside with a free 3 OH group as a co factor + monovalent and divalent cations requirement G-3 OH is compulsory.
SPLICING WITH THE HELP OF SPLICEOSOME: Introns are removed from primary transcriptase in nucleus . exons are ligated to form mRNA molecule- spliceosomes . primary transcript -5 small nuclear RNAs nucleoprotein [SnRNp] complex. RNA segment for the necessary splicing reaction.
RNA EDITING: Changes in RNA nucleotide sequence RNA sequence differs from DNA tem transcribed. RNA editing C-U changes, addition of G or C residues and conversion of U-AMG edits of tRNA and rRNA extremely rare. Mid 1980 in mt genes of Trypanosoma and leishmania. RNA editing mechanism involves addition and deletion of U/C.addition of upto 560 and deletion of up to 41 uridine residue. Addition or deletion small RNA molecule of 40 bp length guide RNA. RNA insertion and deletion at tail region. Self splicing of introns RNA splicing trans esterification helps of guide RNA- aligns by bp itself with the unedited RNA. Splits it into 2 and make new bond between broken at tip of tail of gRNA at 3 end.
RNA editing post transcriptional process radical changes in the AA specified by coding. 1 step: 2 nucleotide of coding sequences. RNA editing plant mitochondria , chloroplast and mammalian nucleus. 2 steps: specificity and biochemical modification. 3 types: Simple editing: conversion of single residue ex: C-U. Insertional editing: insertion of single nucleotides or small runs of nucleotides. Transcriptional strand slipping: ex: G insertion. Pan editing: insertion or deleting of multiple cytidine / uridine residues- help of external antisense guide RNA.