Understanding Drought-Induced Reactive Oxygen Species Generation in Photosynthesis
Leaf responses to drought conditions leading to increased generation of reactive oxygen species (ROS) in photosynthesis are explained. Drought-induced stomatal closure affects CO2 uptake, promoting H2O2 production in peroxisomes and potential ROS production by the photosynthetic electron transport chain. The role of various genes and enzymes in ROS production and redox regulation under drought stress is outlined.
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author. Download presentation by click this link. If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
E N D
Presentation Transcript
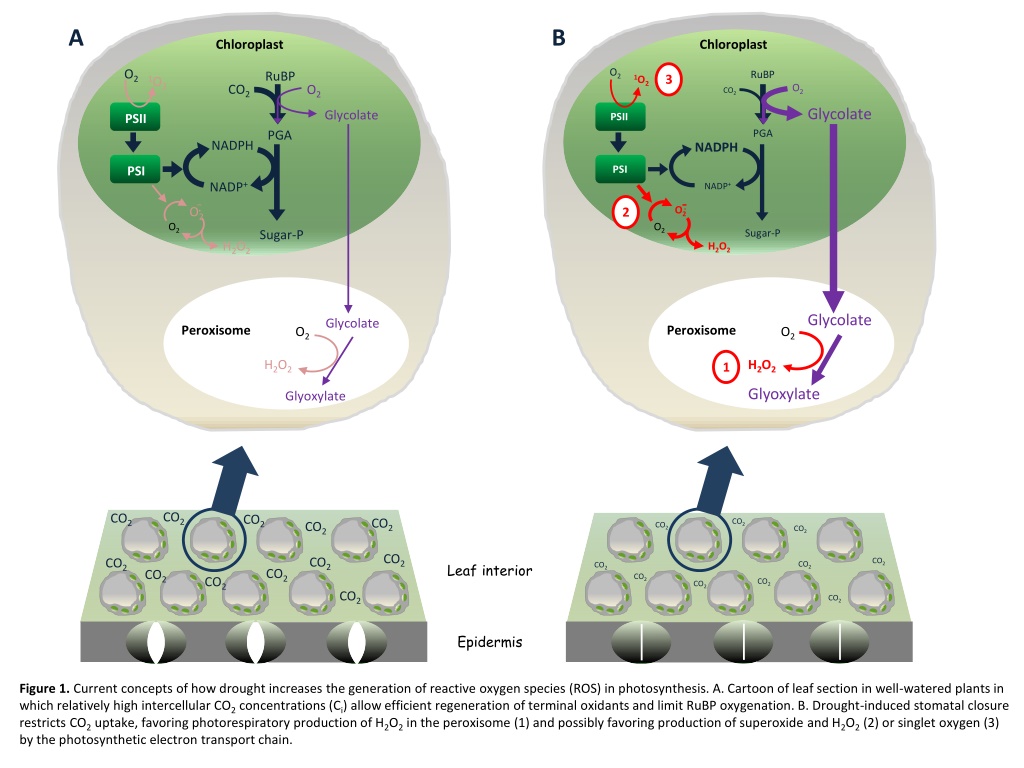
A B Chloroplast Chloroplast O2 O2 RuBP RuBP 3 1O2 1O2 O2 CO2 O2 CO2 Glycolate Glycolate PSII PSII PGA PGA NADPH NADPH PSI PSI NADP+ NADP+ O2. O2. 2 O2 O2 Sugar-P Sugar-P H2O2 H2O2 Glycolate Glycolate Peroxisome Peroxisome O2 O2 H2O2 H2O2 1 Glyoxylate Glyoxylate CO2 CO2 CO2 CO2 CO2 CO2 CO2 CO2 CO2 CO2 CO2 CO2 Leaf interior CO2 CO2 CO2 CO2 CO2 CO2 CO2 CO2 CO2 CO2 Epidermis Figure 1. Current concepts of how drought increases the generation of reactive oxygen species (ROS) in photosynthesis. A. Cartoon of leaf section in well-watered plants in which relatively high intercellular CO2concentrations (Ci) allow efficient regeneration of terminal oxidants and limit RuBP oxygenation. B. Drought-induced stomatal closure restricts CO2uptake, favoring photorespiratory production of H2O2in the peroxisome (1) and possibly favoring production of superoxide and H2O2(2) or singlet oxygen (3) by the photosynthetic electron transport chain.
Plasma Membrane APOPLAST CYTOSOL H2O2 NADPH O2 NADPH oxidase Flavocytochrome Superoxide dismutase O2. - CuZn NADP+ H2O2 O2 Class III peroxidase Superoxide dismutase O2. - Heme CuZn e- O2 O2 Amine oxidase Polyamine oxidase Putrescine Cadaverine Cu FAD H2O2 Spermidine Spermine H2O2 O2 Oxalate oxidase Mn H2O2 Oxalate Figure 2. Multiple ROS-producing enzymes at the cell surface/exterior. Enzymes are shown in blue and their redox cofactors are indicated in yellow. Class III peroxidases may accept electrons from several types of compound to generate superoxide, but in many cases their physiological reductant is not established (reviewed by O Brien et al., 2012).
AGI Gene name Redox class Drought 1 Drought 2 Salt ABA Osmotic At2g29460 At1g17020 At5g35790 At1g06830 At1g35720 At2g28190 At1g08830 At5g17220 At4g02520,At2g02930 At5g06290 At1g56500 At4g39640 At1g78370 At3g03190 At5g18600 GSTU4 glutathione S-transferase TAU4 SRG1 senescence related gene 1 (ascorbate oxidase) G6PD1 glucose-6-phosphate dehydrogenase 1 ROXY6 CC-type glutaredoxin ANNAT1 annexin 1, peroxidase activity CSD2 CuZn superoxide dismutase CSD1 CuZn superoxide dismutase GSTF12 glutathione S-transferase PHI12 GSTF2,GSTF3 glutathione S-transferases PHI2, PHI3 2-cys PrxB 2-cys peroxiredoxin subunit B thioredoxin family protein, suppressor of quenching 1 GGT1 g-glutamyl transpeptidase 1 GSTU20 glutathione S-transferase TAU20 GSTF11 glutathione S-transferase TAU11 ROXY10 CC-type glutaredoxin glutathione ascorbate NADPH glutathione ROS ROS ROS glutathione glutathione thioredoxin thioredoxin glutathione glutathione glutathione glutathione 1.49 1.11 -1.02 -3.24 1.33 -2.72 -1.49 4.76 -1.31 -1.56 -1.05 -1.12 -2.27 -2.21 -1.27 4.36 4.94 -1.83 -1.23 1.80 -1.77 -1.18 3.96 -3.25 -2.13 -1.13 -2.44 -2.29 -1.38 -1.21 1.99 3.92 -1.27 -2.39 1.20 -2.18 -1.36 1.90 -1.55 -1.06 -0.21 -0.64 -0.03 0.53 0.27 3.78 3.47 -2.24 0.41 0.01 -0.15 0.49 0.13 -0.28 -0.48 0.21 -0.24 -0.81 0.99 1.06 2.28 0.60 -0.57 -2.65 0.61 -0.59 0.11 0.52 0.73 -0.12 -0.43 0.02 -0.08 0.18 -0.48 Figure 3. The 15 of the 302 redox-linked genes that respond > 2-fold in the same direction in both drought 1 and drought 2 datasets and their response in related conditions. Data extracted from Genevestigator are shown as log2values compared to controls. Red and green indicate induction and repression. Genes are ordered from top according to the number of conditions in which they respond. The full list of genes and their expression values are given in SupplementalTable S1. For detailsof experiments, see SupplementalTable S2.
Thiol-independent peroxide metabolism Antioxidative systems participate in signaling by their effects on peroxide levels Thiol-dependent peroxide metabolism Signaling is also possible through antioxidative activity per se ROH ROH 2 H2O H2O2 ROOH ROOH ROH ROOH 2 H2O H2O2 APX CAT GST 2CPRX PRXII PRXII 2CPRX SH SH SOH SH SH SOH SH SH ASC GSH H2O2 MDHA GSSG O2 ASC DHA + GSH MDHAR DHAR 2CPRX PRXII S S GSS SH NAD(P)H GSH NAD(P)+ GSSG NTR/TRX NTR/TRX GRX SH SH GRX GSS SH SH S S SH GSH GSSG Dismutation by catalase APX functioning independently of glutathione Ascorbate-glutathione pathway GST glutathione peroxidase function Glutathione-linked peroxiredoxin Thioredoxin-linked peroxiredoxin Figure 4. Peroxide-removing enzymes: roles as antioxidants, in signaling, or both? Cartoon of the best characterized peroxide-metabolizing enzymes in plants. Other mechanisms are possible and for ease of display reactions are not shown stoichiometrically. APX, ascorbate peroxidase. CAT, catalase. DHAR, dehydroascorbate reductase. GRX, glutaredoxin. GST, glutathione S-transferase. MDHAR, monodehydroascorbatereductase. NTR, NADPH-thioredoxinC. PRXII, type II peroxiredoxin. 2CPRX, 2-cys PRX. TRX, thioredoxin.
A B 66 272 37 Total number of drought-induced genes = 375 C - Paraquat: O2, H2O2 (chloroplast) flu: singlet O2 (chloroplast) ABA 246 375 317 External H2O2 Paraquat: O2-, H2O2 flu : singlet O2 cat2: H2O2 cat2: High CO2 ABA (extracellular) (chloroplast) (chloroplast) (peroxisome) DROUGHT 2 DROUGHT 1 129 0 58 Oxidative stress marker transcripts 8 cat2: high CO2 (negative control) cat2: H2O2 (peroxisome) External H2O2 (extracellular) (log2treated/control) 6 4 360 354 372 2 15 21 3 0 Figure 5. Analysis of drought-inducible gene expression in responses to redox perturbation. A. Heatmap of drought-induced genes extracted from Genevestigator and the response of these genes to ABA or oxidative stress (top), and histogram showing expression of oxidative stress marker genes after the different treatments (bottom). Data are shown as log2values compared to Col-0 (wild-type or untreated). Experimental details are given in Supplemental Table S2. Red and green on the heatmap indicate induction and repression according to the color scale shown at the top. The five genes for which data are shown in the bottom histogram are (left to right) APX1, GSTU24, UGT75B1, UGT73B5, and GPX6 (for values, see Supplemental Table S3B). B. Overlap of induced genes (cut-off 2-fold) in the two drought experiments. 375 genes were induced >2-fold in at least one of the experiments (Supplemental Table S3A). C. Number of these 375 genes that were induced > 2-fold by the different oxidative stresses (indicated in red circles within the outer blue circles).