Comprehensive Guide to iCn3D Structure Viewer Features
Explore the diverse functionalities of iCn3D Structure Viewer through this detailed tutorial by Jiyao Wang, emphasizing H-Bonds, Interactions, Electrostatic Potential, Alternate Mutant, Wild Type, VAST+ Alignment, Multiple Chain Alignment, and more. Learn how to utilize different views, graphs, tables, alignments, and annotations effectively for molecular visualization and analysis.
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
You are allowed to download the files provided on this website for personal or commercial use, subject to the condition that they are used lawfully. All files are the property of their respective owners.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.
E N D
Presentation Transcript
Tutorial of iCn3D Structure Viewer - Part 2 Jiyao Wang Staff Scientist NIH/NCBI 5/27/2021 (more details at iCn3D page, more features at gallery)
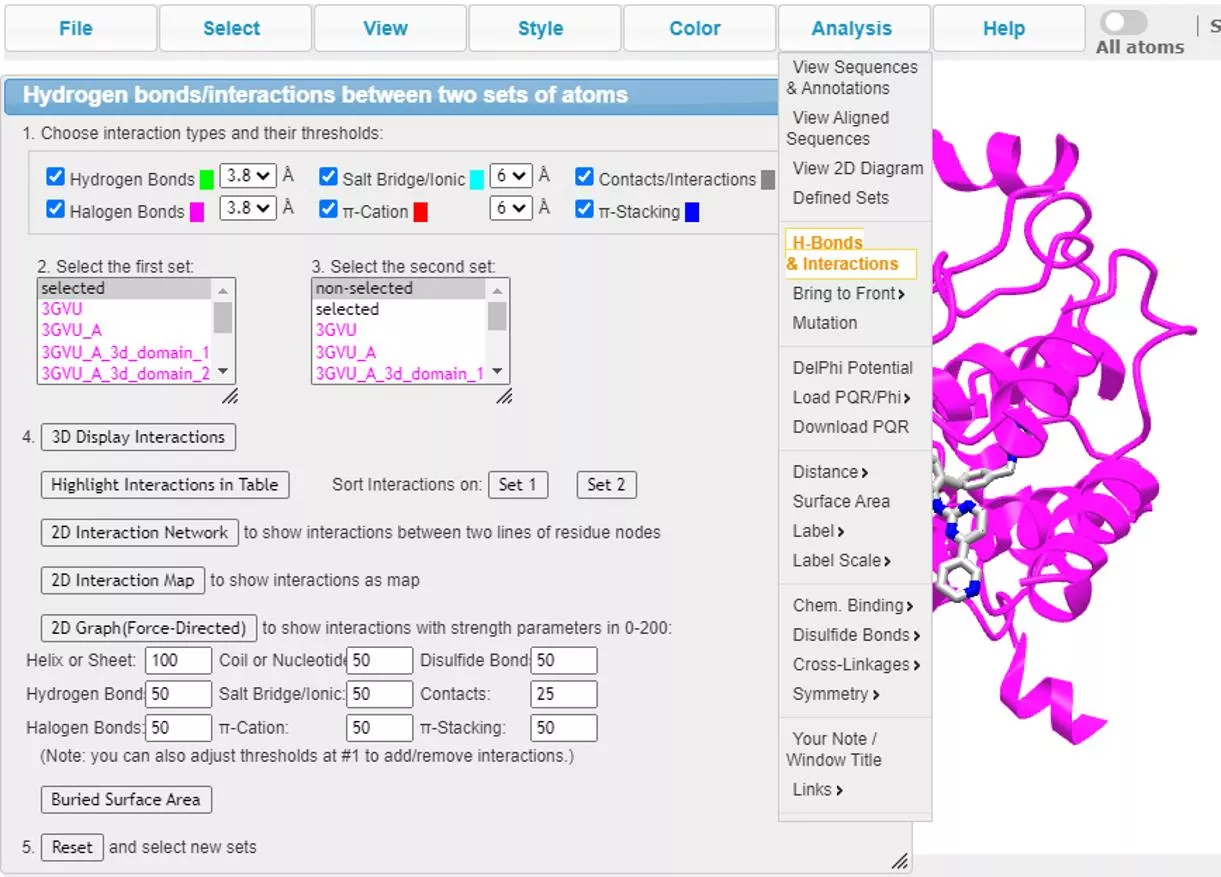
1. H-Bonds & Interactions: 2D Interaction Graph and 2D Interaction Table Share Link: https://structure.ncbi.nlm.nih.gov/icn3d/share.html?Cen4NzadoSLYd2T66
1. H-Bonds & Interactions: Force-directed Graph and No-force Graph Share Link: https://structure.ncbi.nlm.nih.gov/icn3d/share.html?dfjYtCRDnXa4tDpm8
2. Electrostatic Potential Share Link: structure.ncbi.nlm.nih.gov/icn3d/share.html?XCxR6fSTmXHxR3o1A
3. Alternate Mutant and Wild Type Share Link: https://structure.ncbi.nlm.nih.gov/icn3d/share.html?7HCiMKzwTMnCeE2V8
4. VAST+ Alignment: https://www.ncbi.nlm.nih.gov/Structure/vastplus/vastplus.cgi?uid=6m0j
4. VAST+ Alignment: Realign Press letter a to alternate the structures
8. Embed iCn3D using iFrame iFrame: <iframe src="https://www.ncbi.nlm.nih.gov/Structure/icn3d/full.html?mmdbid=1tup&width=300& height=300&showcommand=0&mobilemenu=1&showtitle=0" width="320" height="320" style="border:none"></iframe>
9. Use iCn3D as a widget Example code at: www.ncbi.nlm.nih.gov/Structure/icn3d/example.html add commands (www.ncbi.nlm.nih.gov/Structure/icn3d/icn3d.html#commands) cfg[ command ] = color spectrum ; <script type="text/javascript"> $( document ).ready(function() { function setupViewer(idName, idValue, divid, command) { var cfg = { divid: divid, width: 300, height: 300, mobilemenu: true, showcommand: false, showtitle: false, command: command}; cfg[idName] = idValue; var icn3dui = new icn3d.iCn3DUI(cfg); $.when(icn3dui.show3DStructure()).then(function() { }); } setupViewer( mmdbid', '1tup', 'div0 , color spectrum ); }); </script>
10. Use iCn3D to write Node.js script icn3d package at npm: www.npmjs.com/package/icn3d Example Node.js script: github.com/ncbi/icn3d/blob/master/icn3dnode/interaction2.js Installation Node.js Script npm install icn3d global.THREE = require('three'); let jsdom = require('jsdom'); global.$ = require('jquery')(new jsdom.JSDOM().window); #dependency: npm install jquery npm install jsdom npm install three let icn3d = require('icn3d'); let icn3dui = new icn3d.iCn3DUI({});
11. Build iCn3D install nodejs and npm. git clone https://github.com/ncbi/icn3d.git npm install -g gulp npm install gulp Complete package of iCn3D including Three.js and jQuery can be downloaded from https://www.ncbi.nlm.nih.gov/Structure/icn3d/icn3d-3.1.3.zip. full.html:
12. iCn3D Development Instruction: www.ncbi.nlm.nih.gov/Structure/icn3d/icn3d.html#addclass Download iCn3D local copy: www.ncbi.nlm.nih.gov/Structure/icn3d/icn3d-3.1.3.zip Example: module.html loadStateFile.js <script type="module"> import * as icn3d from './icn3d.module.js'; // add new classes import {LoadStateFile} from './loadStateFile.js ; // import any classes from icn3d.module.js import {LoadScript} from './icn3d.module.js'; // class name starts with an upper-case letter class LoadStateFile { // pass the instance of the class iCn3D constructor(icn3d) { this.icn3d = icn3d; } var icn3dui = new icn3d.iCn3DUI(cfg); $.when(icn3dui.show3DStructure()).then(function() { // call classes here var loadStateFileCls = new LoadStateFile(icn3dui.icn3d); loadStateFileCls.loadStateFile('color spectrum'); }); // functions start with a lower-case letter // use "ic" to access the instance of iCn3D class loadStateFile(fileStr) { var ic = this.icn3d; // "ic" has a lot of class instances such as "loadScriptCls" ic.loadScriptCls.loadScript(fileStr, true); } } // export your class export {LoadStateFile}