RNA Secondary Structure Prediction in Bioinformatics
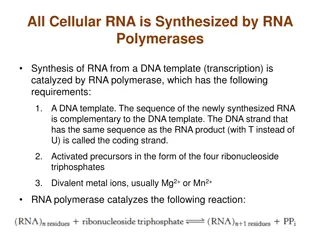
RNA secondary structure prediction is a key concept in bioinformatics, encompassing features like stems, loops, and bulges. This presentation delves into the importance of RNA beyond mRNA, highlighting rRNA, tRNA, and regulatory RNA roles. The canonical base pairs A-U and C-G shape the single-stranded RNA folding, with base-paired structures defining its secondary structure. Visual aids detail the secondary structures of tRNA, ribosomal RNA, and 6S RNA. The lecture also covers common secondary structure features such as bulges, hairpins, and internal loops in RNA molecules.
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
You are allowed to download the files provided on this website for personal or commercial use, subject to the condition that they are used lawfully. All files are the property of their respective owners.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.
E N D
Presentation Transcript
RNA Secondary Structure Prediction BMI/CS 776 www.biostat.wisc.edu/bmi776/ Spring 2018 Anthony Gitter gitter@biostat.wisc.edu These slides, excluding third-party material, are licensed under CC BY-NC 4.0 by Mark Craven, Colin Dewey, and Anthony Gitter
Goals for Lecture Key concepts RNA secondary structure Secondary structure features: stems, loops, bulges Pseudoknots Nussinov algorithm Adapting Nussinov to take free energy into account 2
Why RNA is Interesting Messenger RNA (mRNA) isn t the only important class of RNA ribosomal RNA (rRNA) ribosomes are complexes that incorporate several RNA subunits in addition to numerous protein units transfer RNA (tRNA) transport amino acids to the ribosome during translation the spliceosome, which performs intron splicing, is a complex with several RNA units microRNAs and others that play regulatory roles many viruses (e.g. HIV) have RNA genomes guide RNA sequence complementary determines whether to cleave DNA Folding of an mRNA can be involved in regulating the gene s expression 3
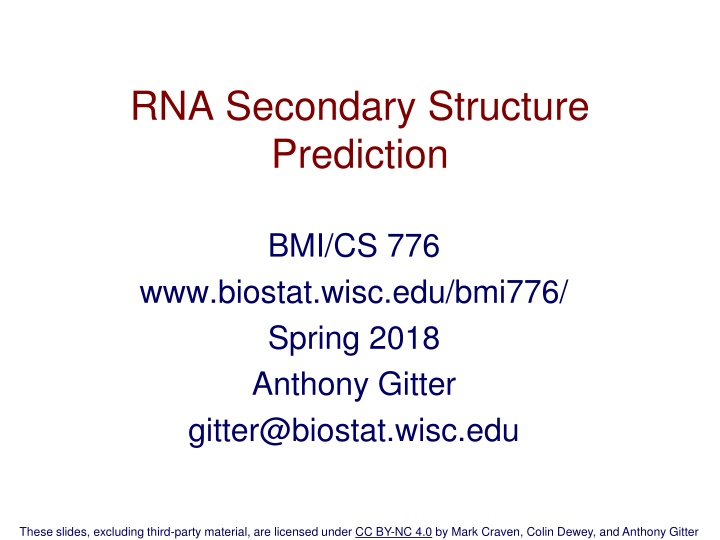
RNA Secondary Structure RNA is typically single stranded Folding, in large part is determined by base-pairing A-U and C-G are the canonical base pairs other bases will sometimes pair, especially G-U Base-paired structure is referred to as the secondary structure of RNA Related RNAs often have homologous secondary structure without significant sequence similarity 4
tRNA Secondary Structure Scitable tertiary structure 5
Small Subunit Ribosomal RNA Secondary Structure 6
Secondary Structure Features bulge hairpin loop stem internal loop 8
Four Key Problems Predicting RNA secondary structure Given: RNA sequence Do: predict secondary structure that sequence will fold into Focus for today Searching for instances of a given structure Given: an RNA sequence or its secondary structure Do: find sequences that will fold into a similar structure Modeling a family of RNAs Given: a set of RNA sequences with similar secondary structure Do: construct a model that captures the secondary structure regularities of the set Identifying novel RNA genes Given: a pair of homologous DNA sequences Do: identify subsequences that appear to have highly conserved RNA secondary structure (putative RNA genes) 9
RNA Folding Assumption Algorithms we ll consider assume that base pairings do not cross For base-paired positions i, i and j, j , with i < i and j < j , we must have either i < i < j < j or j < j < i < i (not nested) i i j j i < j < j < i or j < i < i < j (nested) i j j i Can t have i < j < i < j or j < i < j < i 10
Pseudoknots pseudoknot These crossings are called pseudoknots Dynamic programming breaks down if pseudoknots are allowed Fortunately, they are not very frequent Modern software does support them Akiyama et al. 2018 Figure from Seliverstov et al. BMC Microbiology, 2005 11
Simplest RNA Secondary Structure Task Given: An RNA sequence The constraint that pseudoknots are not allowed Do: Find a secondary structure for the RNA that maximizes the number of base pairing positions 12
Predicting RNA Secondary Structure: the Nussinov Algorithm [Nussinov et al., SIAM Journal of Applied Mathematics 1978] Key idea: Do this using dynamic programming start with small subsequences progressively work to larger ones 13
DP in the Nussinov Algorithm j G G G A A A U C C i G G G A A A U C C max # of paired bases in subsequence [i, j] Figure 10.8 from textbook 14
DP in the Nussinov Algorithm 1 if and are complementary i j x x = Let ( , ) i j 0 otherwise Initialization: , ( = = ) 1 = 0 for to 2 i i i L , ( = ) 0 for to 1 i i i L Recursion + ( , 1 ) i j , ( i ) 1 - j i , ( i = ) max j + + ( , 1 ) 1 - k , ( k ) j i j + + max k i , ( i ) ( , 1 ) j max # of paired bases in subsequence [i, j] j 15
Nussinov Algorithm Traceback push 1 ( onto ) stack ,L repeat until i,j stack empty is pop ( ) if continue i + i j = + else if ( , 1 j + ) , ( i push ) ( , 1 j ) j j i j = ) 1 else if , ( i ) 1 j , ( i + push ) j , ( i i j ) 1 , ( i = , ( i else if ( , 1 i, j + ) ) j record i = base j + pair ) 1 push ( , 1 i + , ( i + , 1 + = , ( i else for to 1 , j if : 1 j- ) ( ) ) k k k j j push ( 1 ) k push ( ) i,k break 16
Predicting RNA Secondary Structure by Energy Minimization It s na ve to predict folding just by maximizing the number of base pairs However, we can generalize the key recurrence relation so that we re minimizing free energy instead + ( 1 j- ) E i j , ( ) 1 E E i, = , ( i E ) min j + + min k i ( ) ( 1 j , ) i, k E k i j , ( P ) j case that i and j are base paired 17
Predicting RNA Secondary Structure by Energy Minimization A sophisticated program, such as Mfold [Zuker et al.], can take into account free energy of the local environment of [i, j] + , ( , ( i, ) LoopEnergy ( ) 1 j i j j i + ( i, + + + ) 1 ( ) StackingEn j ergy , 1 P ) 1 k ( 1 j , ) 1 j i i j , P i + + + + min k ) BulgeEnerg y( ) ( 1 k i j , 1 = , ( i P ) min j + + + min k ( ) BulgeEnerg y( ) ( 1 j , ) 1 i, j k P i k 1 + + + + + min , l k ( ) LoopEnergy ( ) ( 1 j , ) 1 i, j k l P i k l 1 + + + min k j ( ) ( 1 k , + ) ( , 1 ) 1 i, j E i E k j i 18
Predicting RNA Secondary Structure by Energy Minimization ) 1 + ) 1 + + + + + , ( i ) LoopEnergy ( j j i min , l k ( ) LoopEnergy ( ) ( 1 j , i, j k l P i k l 1 c a g u c i+k+1 j-l-1 c u u u c g i j c c a u c c a g u c j i ) 1 + + + + min k ( ) BulgeEnerg y( ) ( 1 i, j k P i k j , 1 a g u c i+k+1 + + ) 1 + + ) 1 j-1 ( ) StackingEn ergy , ( i , 1 j , ( 1 j , i, j j i P i u g c c c g i+1 j-1 c a g u c j i c g i j a u 19
Mfold example GGGAAAUCC G = -0.80 kcal/mol G = 0.20 kcal/mol http://unafold.rna.albany.edu/ 20