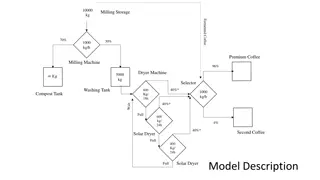
Workflow Management and Systematic Organization
A workflow comprises orchestrated business activities organized into processes that transform materials, provide services, or process information. Learn about different types of workflows, examples, the Kepler system, CONNJUR Workflow Builder, provenance types, and the importance of reproducibility in scientific workflows.
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
You are allowed to download the files provided on this website for personal or commercial use, subject to the condition that they are used lawfully. All files are the property of their respective owners.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.
E N D
Presentation Transcript
Introduction to CONNJUR Workflow Builder and Yes Workflow 2017 Summer Workshop: June 29, 2017
Workflows (Wikipedia) A workflow consists of an orchestrated and repeatable pattern of business activity enabled by the systematic organization of resources into processes that transform materials, provide services, or process information. It can be depicted as a sequence of operations, declared as work of a person or group, an organization of staff, or one or more simple or complex mechanisms. From a more abstract or higher-level perspective, workflow may be considered a view or representation of real work. The flow being described may refer to a document, service or product that is being transferred from one step to another.
Workflows (Examples) On the first day Frank described an iterative workflow by which a spectroscopist converts Varian/Bruker data into nmrPipe format, resolves ambiguities, performs preliminary processing, resolves phasing, reprocesses, iterate until done. Bertram Ludascher: ASAP Automate computation Scalable Adaptable for reuse Provenance: capture processing history and data lineage
Kepler Lud scher, Bertram, Ilkay Altintas, Chad Berkley, Dan Higgins, Efrat Jaeger, Matthew Jones, Edward A Lee, Jing Tao, and Yang Zhao. 2006. Scientific Workflow Management and the Kepler System. Concurrency and Computation: Practice and Experience 18 (10): 1039 65.
CONNJUR Workflow Builder M. Fenwick, G. Weatherby, J. Vyas, C. Sesanker, T. O. Martyn, H. J. Ellis, and M. R. Gryk, (2015) CONNJUR Workflow Builder: a software integration environment for spectral reconstruction. J Biomol NMR,62, 313-26.
Provenance Types (Michael Wilde, Argonne Labs) Prospective Provenance: the specification of the workflows procedure calls and data dependencies (acqu, workflow) Retrospective Provenance: the recordings of when and where each procedure ran, and how each invocation behaved (acqus, reconstruction)
Yes Workflow Annotation system for Prospective Provenance from scripts
Reproducibility Replicability vs. reproducibility or is it the other way around? Reproducibility, replicability, reusability, repeatability
Reproducibility (Dagstuhl Working Group) PRIMAD Platform Portability vs. reproducibility (OS or hardware platforms) Research Objective (goal of computation) Implementation (Fast Fourier Transform) Method (Fourier Transform) Actors (Dagstuhl group defines agent as human) Data (data used in study) Rauber, A., Braganholo, V., Dittrich, J., et al. (2016). PRIMAD Information gained by different types of reproducibility. In Reproducibility of Data-Oriented Experiments in e-Science (Dagstuhl Seminar 16041). Friere, J., Fuhr, N., & Rauber, A. editors. Gryk, M. & Lud scher, B. (2017). Workflows and provenance: Towards information science solutions for the natural sciences. Library Trends, in press.
Metadata Definition Definition 1: Data about Data
Workflow -> Provenance -> Reproducibility All rely on the capture of metadata Definition 1: Data about Data
Workflow -> Provenance -> Reproducibility All rely on the capture of metadata Definition 1: Data about Data Definition 2: Metadata As Surrogate
Workflow -> Provenance -> Reproducibility All rely on the capture of metadata Definition 1: Data about Data Definition 2: Metadata As Surrogate
Yes Workflow #@BEGIN main #@IN raw_turkey #@OUT cooked_turkey #@BEGIN survey_guests #@OUT food_allergies #@END survey_guests #@BEGIN brining #@IN raw_turkey @URI store:shnucks_turkey #@PARAM seasonings #@OUT brined_turkey #@END brining #@BEGIN weighing #@IN brined_turkey #@OUT weighed_turkey #@OUT weight #@END weighing #@BEGIN stuffing #@IN weighed_turkey #@IN stuffing_ingredients #@IN food_allergies #@OUT stuffed_turkey #@END stuffing #@BEGIN baking #@PARAM weight #@PARAM temperature #@PARAM duration #@IN stuffed_turkey #@OUT cooked_turkey @URI plate:delicious_turkey #@END baking #@END main @URI store:shnucks_turkey @URI plate:delicious_turkey
Yes Workflow echo 'Converting from Varian to NMRPipe format' var2pipe -in ./fid \ -xN 1024 -yN -xT 512 -yT -xMODE Complex -xSW 12000 -ySW -xOBS 599.5694 -yOBS -xCAR 4.772 -yCAR -xLAB 1H -yLAB -ndim 3 -aq2D -out ./data/hnco%03d.pipe -verb -ov 128 64 -yMODE 6000 -zN 64 \ -zT 32 \ States-TPPI -zSW 2000 \ 125.768 -zCAR 119 \ 13C -zLAB 15N \ States \ -zMODE Rance-Kay \ -zOBS 60.7438 \ 45 sleep 1 echo 'Transforming 3-dimensional NMR data!' echo 'Processing F3/F1 dimensions first' xyz2pipe -in data/hnco%03d.pipe -x -verb \ | nmrPipe -fn SOL -mode 1 -fl 16 -fs 1 -poly \ | nmrPipe -fn CBF -last 12 \ | nmrPipe -fn SP -off 0.39 -end 0.98 -pow 2 -size 512 -c 0.5 \ | nmrPipe -fn ZF -size 2048 \ | nmrPipe -fn FT -verb \ | nmrPipe -fn PS -p0 0 -p1 0 -di \ | nmrPipe -fn EXT -left -sw -verb \ | nmrPipe -fn TP \ | nmrPipe -fn SP -off 0.39 -end 0.98 -pow 2 -size 64 -c 0.5 \ | nmrPipe -fn ZF -size 256 \ | nmrPipe -fn FT -verb \ | nmrPipe -fn PS -p0 0 -p1 0 -di \ | pipe2xyz -out ft/hnco%03d.ft2 y xyz2pipe -in ft/hnco%03d.ft2 -z -verb \ | nmrPipe -fn SP -off 0.39 -end 0.98 -pow 2 -size 32 -c 0.5 \ | nmrPipe -fn ZF -size 128 \ | nmrPipe -fn FT -neg -verb \ | nmrPipe -fn PS -p0 0 -p1 0 -di \ | pipe2xyz -out ft/hnco%03d.ft3 -z Website
bash vs. csh the shell wars #! /bin/csh # = comment which nmrPipe tool? nmrPipe, var2pipe, xyz2pipe, etc. # My Processing Script nmrPipe -in mydata.pipe \ | nmrPipe -fn SOL -mode 1 -fl 16 -fs 1 -poly \ | nmrPipe -fn CBF -last 12 \ | nmrPipe -fn GMB -lb -7 -gb 0.1 -size 512 -c 0.5 \ | nmrPipe -fn ZF -size 2048 \ | nmrPipe -fn FT -verb \ | nmrPipe -fn PS -p0 68.6 -p1 -34.8 -di \ | nmrPipe -fn EXT -left -sw -verb \ | nmrPipe -fn TP \ | nmrPipe -fn LP -fb -ord 30 -x1 2 -xn 128 -pred 64 -fix -fixMode 1 -after \ | nmrPipe -fn SP -off 0.39 -end 0.98 -pow 2 -size 192 -c 0.5 \ | nmrPipe -fn ZF -size 256 \ | nmrPipe -fn FT -verb \ | nmrPipe -fn PS -p0 -9.0 -p1 20.0 -di \ -out mydata.ft2 -ov watch for trailing spaces! Syntax? filenames and filesystems How much of the above information is NMR related? How much is related to nmrPipe or the computer we are using? 16
Functionality and Function Order! #! /bin/csh # My Processing Script nmrPipe -in mydata.pipe \ | nmrPipe -fn SOL -mode 1 -fl 16 -fs 1 -poly \ | nmrPipe -fn CBF -last 12 \ | nmrPipe -fn GMB -lb -7 -gb 0.1 -size 512 -c 0.5 \ | nmrPipe -fn ZF -size 2048 \ | nmrPipe -fn FT -verb \ | nmrPipe -fn PS -p0 68.6 -p1 -34.8 -di \ | nmrPipe -fn EXT -left -sw -verb \ | nmrPipe -fn TP \ | nmrPipe -fn LP -fb -ord 30 -x1 2 -xn 128 -pred 64 -fix -fixMode 1 -after \ | nmrPipe -fn SP -off 0.39 -end 0.98 -pow 2 -size 192 -c 0.5 \ | nmrPipe -fn ZF -size 256 \ | nmrPipe -fn FT -verb \ | nmrPipe -fn PS -p0 -9.0 -p1 20.0 -di \ -out mydata.ft2 -ov What procedures do we use to massage our data? What procedures do we use to transform our data? 17