Understanding EMBOSS Needle: Pairwise Sequence Alignment Tool
EMBOSS Needle is a pairwise sequence alignment tool that uses the Needleman-Wunsch algorithm to find the optimal global alignment between two input sequences. It is available online through EMBOSS and requires entering two protein/DNA sequences of the same length to generate alignment results, including gaps. The tool can be accessed via Web Form, REST API, and supports various alignment modes such as global, local, and pairwise sequence alignment. Parameters like sequence input, alignment options, and job submission play a vital role in obtaining alignment outcomes.
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author. Download presentation by click this link. If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
E N D
Presentation Transcript
Needle Needle
Content Content Introduction Objectives Input & Output Algorithms & Alignments Parameters Conclusion
Introductio Introductio n n EMBOSS Needle reads two input sequences and writes their optimal global sequence alignment to file.
Needle is a pairwise alignment tool designed to write the optimal global alignment. Needle is sub-software/tools available in EMBOSS.
Objectives Objectives Backend Backend Algorithm Algorithm It uses the Needleman-Wunsch alignment algorithm to find the optimum alignment. Analyzes Analyzes The tool writes optimal global sequence alignment for sequences and finds optimal alignment including gaps. Available on Available on Needle is available in EMBOSS as an online tool for pairwise alignment.
Input & Output Input & Output Input Input Output Output Needle requires to enter or upload file of two sequences (Protein/DNA) o same lengths. Needle finds the optimal alignment for both the input sequences (global sequence alignment).
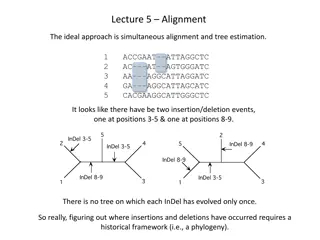
Needleman Needleman - -Wunsch Wunsch Optimal matching algorithm used in bioinformatics. Access Access Needle can be accessed via Web Form REST API Algorithm Algorithm & & Alignment Alignment s s Global Global Alignment Alignment End to end sequence alignment takes place. MSA MSA Needle uses MSA algorithms to finds optimal alignments. Local Local Alignment Alignment Query is matched with the portion of your reference sequence PSA PSA Pairwise sequence alignment-Finds similar regions between two biological sequences.
Needleman- Wunsch-Pairwise sequence alignment.
Parameters Parameters Input Input Sequence Sequence Requires to enter two protein sequences to perform global alignment 1st 2nd Options Options Requires to set the options for pairwise sequence alignment Submissio Submissio n n 3rd Requires to submit the job to get the alignment results
Alignment results by EMBOSS Needle for FASTA sequences of cycling DOF factor 1 [Arabidopsis thaliana] as first protein and dof zinc finger protein [Zea mays] as the second protein sequence.
Conclusion Conclusion Performing Global alignment for query protein sequences through EMBOSS Needle is efficient. It uses the Needleman-Wunsch alignment algorithm to find the optimum alignment (including gaps) of two sequences along their entire length. It forms the matrix and track the optimum sequence.