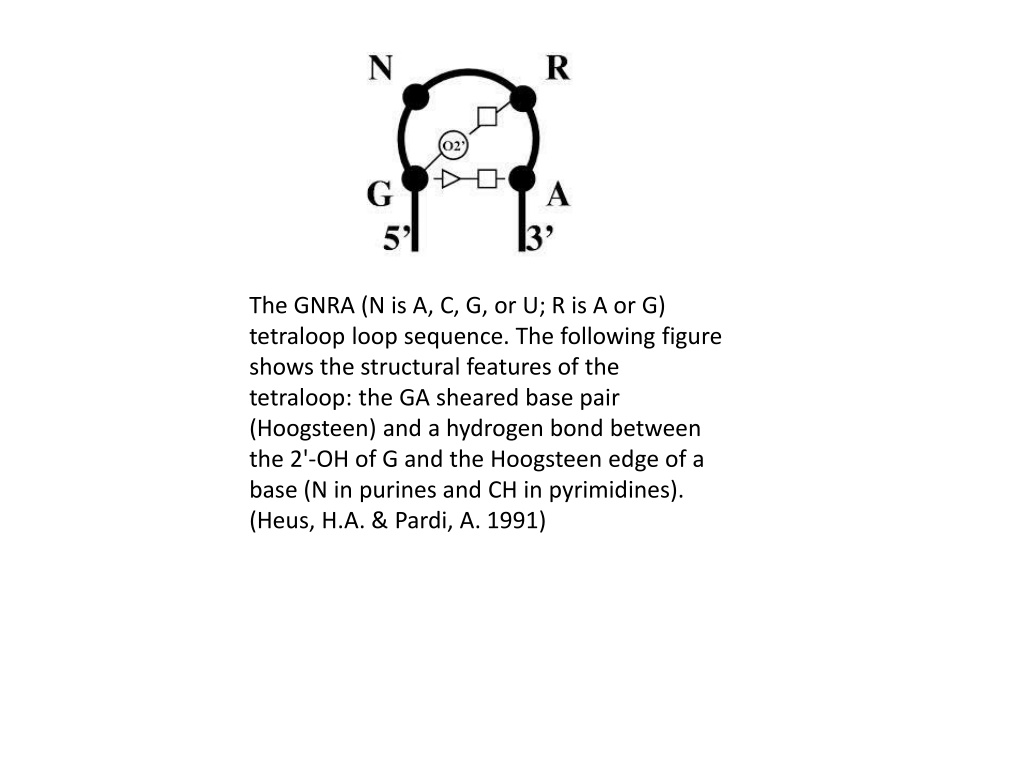
Structural Features of GNRA Tetraloop in DNA
The GNRA tetraloop in DNA consists of a sheared base pair (Hoogsteen) and hydrogen bonding interactions, essential for its structural stability. This loop sequence exhibits specific structural characteristics, such as the interaction between the 2'-OH of G and the Hoogsteen edge of a base. Chemical structures depict the Watson-Crick and Hoogsteen base pairs, with the Hoogsteen geometry achievable by purine rotation and base-flipping. DNA base flipping is a crucial mechanism involved in various biological processes.
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
You are allowed to download the files provided on this website for personal or commercial use, subject to the condition that they are used lawfully. All files are the property of their respective owners.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.
E N D
Presentation Transcript
The GNRA (N is A, C, G, or U; R is A or G) tetraloop loop sequence. The following figure shows the structural features of the tetraloop: the GA sheared base pair (Hoogsteen) and a hydrogen bond between the 2'-OH of G and the Hoogsteen edge of a base (N in purines and CH in pyrimidines). (Heus, H.A. & Pardi, A. 1991)
Chemical structures for Watson-Crick and Hoogsteen A T and G C+ base pairs. The Hoogsteen geometry can be achieved by purine rotation around the glycosidic bond ( ) and base-flipping ( ; rotating 180 degrees out of the helix), affecting simultaneously C8 and C1 (yellow).[1]
An external file that holds a picture, illustration, etc. Object name is gkf48101.jpg An external file that holds a picture, illustration, etc. Object name is gkf481t2.jpg