Mus musculus Cytochrome b Gene Sequence
"Explore the complete nucleotide sequence of the Mus musculus cytochrome b gene, isolated from RF-4 strain. The sequence is aligned and presented in the FASTA format, providing insights into the genetic composition of this mitochondrial gene in the house mouse."
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author. If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
You are allowed to download the files provided on this website for personal or commercial use, subject to the condition that they are used lawfully. All files are the property of their respective owners.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.
E N D
Presentation Transcript
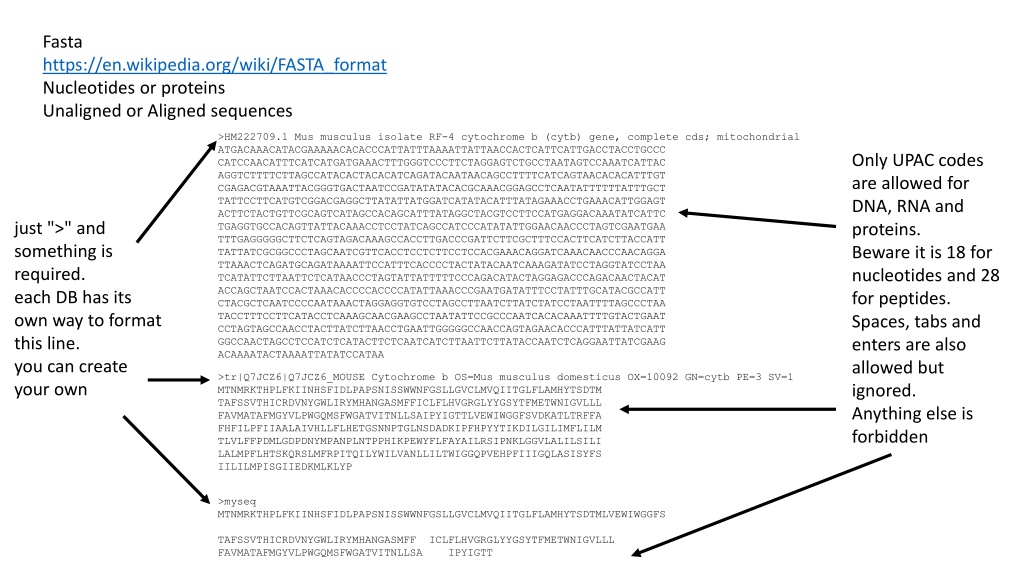
Fasta https://en.wikipedia.org/wiki/FASTA_format Nucleotides or proteins Unaligned or Aligned sequences >HM222709.1 Mus musculus isolate RF-4 cytochrome b (cytb) gene, complete cds; mitochondrial ATGACAAACATACGAAAAACACACCCATTATTTAAAATTATTAACCACTCATTCATTGACCTACCTGCCC CATCCAACATTTCATCATGATGAAACTTTGGGTCCCTTCTAGGAGTCTGCCTAATAGTCCAAATCATTAC AGGTCTTTTCTTAGCCATACACTACACATCAGATACAATAACAGCCTTTTCATCAGTAACACACATTTGT CGAGACGTAAATTACGGGTGACTAATCCGATATATACACGCAAACGGAGCCTCAATATTTTTTATTTGCT TATTCCTTCATGTCGGACGAGGCTTATATTATGGATCATATACATTTATAGAAACCTGAAACATTGGAGT ACTTCTACTGTTCGCAGTCATAGCCACAGCATTTATAGGCTACGTCCTTCCATGAGGACAAATATCATTC TGAGGTGCCACAGTTATTACAAACCTCCTATCAGCCATCCCATATATTGGAACAACCCTAGTCGAATGAA TTTGAGGGGGCTTCTCAGTAGACAAAGCCACCTTGACCCGATTCTTCGCTTTCCACTTCATCTTACCATT TATTATCGCGGCCCTAGCAATCGTTCACCTCCTCTTCCTCCACGAAACAGGATCAAACAACCCAACAGGA TTAAACTCAGATGCAGATAAAATTCCATTTCACCCCTACTATACAATCAAAGATATCCTAGGTATCCTAA TCATATTCTTAATTCTCATAACCCTAGTATTATTTTTCCCAGACATACTAGGAGACCCAGACAACTACAT ACCAGCTAATCCACTAAACACCCCACCCCATATTAAACCCGAATGATATTTCCTATTTGCATACGCCATT CTACGCTCAATCCCCAATAAACTAGGAGGTGTCCTAGCCTTAATCTTATCTATCCTAATTTTAGCCCTAA TACCTTTCCTTCATACCTCAAAGCAACGAAGCCTAATATTCCGCCCAATCACACAAATTTTGTACTGAAT CCTAGTAGCCAACCTACTTATCTTAACCTGAATTGGGGGCCAACCAGTAGAACACCCATTTATTATCATT GGCCAACTAGCCTCCATCTCATACTTCTCAATCATCTTAATTCTTATACCAATCTCAGGAATTATCGAAG ACAAAATACTAAAATTATATCCATAA Only UPAC codes are allowed for DNA, RNA and proteins. Beware it is 18 for nucleotides and 28 for peptides. Spaces, tabs and enters are also allowed but ignored. Anything else is forbidden just ">" and something is required. each DB has its own way to format this line. you can create your own >tr|Q7JCZ6|Q7JCZ6_MOUSE Cytochrome b OS=Mus musculus domesticus OX=10092 GN=cytb PE=3 SV=1 MTNMRKTHPLFKIINHSFIDLPAPSNISSWWNFGSLLGVCLMVQIITGLFLAMHYTSDTM TAFSSVTHICRDVNYGWLIRYMHANGASMFFICLFLHVGRGLYYGSYTFMETWNIGVLLL FAVMATAFMGYVLPWGQMSFWGATVITNLLSAIPYIGTTLVEWIWGGFSVDKATLTRFFA FHFILPFIIAALAIVHLLFLHETGSNNPTGLNSDADKIPFHPYYTIKDILGILIMFLILM TLVLFFPDMLGDPDNYMPANPLNTPPHIKPEWYFLFAYAILRSIPNKLGGVLALILSILI LALMPFLHTSKQRSLMFRPITQILYWILVANLLILTWIGGQPVEHPFIIIGQLASISYFS IILILMPISGIIEDKMLKLYP >myseq MTNMRKTHPLFKIINHSFIDLPAPSNISSWWNFGSLLGVCLMVQIITGLFLAMHYTSDTMLVEWIWGGFS TAFSSVTHICRDVNYGWLIRYMHANGASMFF ICLFLHVGRGLYYGSYTFMETWNIGVLLL FAVMATAFMGYVLPWGQMSFWGATVITNLLSA IPYIGTT
>HM222709.1 Mus musculus isolate RF-4 cytochrome b (cytb) gene, complete cds; mitochondrial ATGACAAACATACGAAAAACACACCCATTATTTAAAATTATTAACCACTCATTCATTGACCTACCTGCCC CATCCAACATTTCATCATGATGAAACTTTGGGTCCCTTCTAGGAGTCTGCCTAATAGTCCAAATCATTAC AGGTCTTTTCTTAGCCATACACTACACATCAGATACAATAACAGCCTTTTCATCAGTAACACACATTTGT CGAGACGTAAATTACGGGTGACTAATCCGATATATACACGCAAACGGAGCCTCAATATTTTTTATTTGCT TATTCCTTCATGTCGGACGAGGCTTATATTATGGATCATATACATTTATAGAAACCTGAAACATTGGAGT ACTTCTACTGTTCGCAGTCATAGCCACAGCATTTATAGGCTACGTCCTTCCATGAGGACAAATATCATTC TGAGGTGCCACAGTTATTACAAACCTCCTATCAGCCATCCCATATATTGGAACAACCCTAGTCGAATGAA TTTGAGGGGGCTTCTCAGTAGACAAAGCCACCTTGACCCGATTCTTCGCTTTCCACTTCATCTTACCATT TATTATCGCGGCCCTAGCAATCGTTCACCTCCTCTTCCTCCACGAAACAGGATCAAACAACCCAACAGGA TTAAACTCAGATGCAGATAAAATTCCATTTCACCCCTACTATACAATCAAAGATATCCTAGGTATCCTAA TCATATTCTTAATTCTCATAACCCTAGTATTATTTTTCCCAGACATACTAGGAGACCCAGACAACTACAT ACCAGCTAATCCACTAAACACCCCACCCCATATTAAACCCGAATGATATTTCCTATTTGCATACGCCATT CTACGCTCAATCCCCAATAAACTAGGAGGTGTCCTAGCCTTAATCTTATCTATCCTAATTTTAGCCCTAA TACCTTTCCTTCATACCTCAAAGCAACGAAGCCTAATATTCCGCCCAATCACACAAATTTTGTACTGAAT CCTAGTAGCCAACCTACTTATCTTAACCTGAATTGGGGGCCAACCAGTAGAACACCCATTTATTATCATT GGCCAACTAGCCTCCATCTCATACTTCTCAATCATCTTAATTCTTATACCAATCTCAGGAATTATCGAAG ACAAAATACTAAAATTATATCCATAA >HE647856.1 Homo sapiens mitochondrial cytb gene for cytochrome b ATGACCCCAATACGCAAAACTAACCCCCTAATAAAATTAATTAACCACTCATTCATCGACCTCCCCACCC CATCCAACATCTCCGCATGATGAAACTTCGGCTCACTCCTTGGCGCCTGCCTGATCCTCCAAATCACCAC AGGACTATTCCTAGCCATGCACTACTCACCAGACGCCTCAACCGCCTTTTCATCAATCGCCCACATCACT CGAGACGTAAATTATGGCTGAATCATCCGCTACCTTCACGCCAATGGCGCCTCAATATTCTTTATCTGCC TCTTCCTACACATCGGGCGAGGCCTATATTACGGATCATTTCTCTACTCAGAAACCTGAAACATCGGCAT TATCCTCCTGCTTGCAACTATAGCAACAGCCTTCATAGGCTATGTCCTCCCGTGAGGCCAAATATCATTC TGAGGGGCCACAGTAATTACAAACTTACTATCCGCCATCCCATACATTGGGACAGACCTAGTTCAATGAA TCTGAGGAGGCTACTCAGTAGACAGTCCCACCCTCACACGATTCTTTACCTTTCACTTCATCTTGCCCTT CATTATTGCAGCCCTAGCAACACTCCACCTCCTATTCTTGCACGAAACGGGATCAAACAACCCCCTAGGA ATCACCTCCCATTCCGATAAAATCACCTTCCACCCTTACTACACAATCAAAGACGCCCTCGGCTTACTTC TCTTCCTTCTCTCCTTAATGACATTAACACTATTCTCACCAGACCTCCTAGGCGACCCAGACAATTATAC CCTAGCCAACCCCTTAAACACCCCTCCCCACATCAAGCCCGAATGATATTTCCTATTCGCCTACACAATT CTCCGATCCGTCCCTAACAAACTAGGAGGCGTCCTTGCCCTATTACTATCCATCCTCATCCTAGCAATAA TCCCCATCCTCCATATATCCAAACAACAAAGCATAATATTTCGCCCACTAAGCCAATCACTTTATTGACT CCTAGCCGCAGACCTCCTCATTCTAACCTGAATCGGAGGACAACCAGTAAGCTACCCTTTTACCATCATT GGACAAGTAGCATCCGTACTATACTTCACAACAATCCTAATCCTAATACCAACTATCTCCCTAATTGAAA ACAAAATACTCAAATGGGCCT Fasta: multiple sequences are allowed in the same file
One line starting with a ">" (greater-than) sign, followed by a two-letter code describing the sequence type (P1, F1, DL, DC, RL, RC, or XX), followed by a semicolon, followed by the sequence identification code. One line containing a textual description of the sequence. One or more lines containing the sequence itself. When multiple chains are included in a single sequence a "/" (slash) character is used as a delimiter The end of the sequence is marked by a "*" (asterisk) character. PIR format AKA NBRF https://www.bioinformatics.nl/tools/crab_pir.html Nucleotides or proteins Unaligned or Aligned sequences >P1;T22GFP sequence:T22GFP:::::::0.00:0.00 MRRWCYRKCYKGYCYRKCRGGSSRSSSKGEELFTGVVPILVELDGDVNGHKFSVSGEGEGDATYGKLTLKFI CTTGKLPVPWPTLVTTLTYGVQCFSRYPDHMKRHDFFKSAMPEGYVQERTISFKDDGNYKTRAEVKFEGDTL VNRIELKGIDFKEDGNILGHKLEYNYNSHNVYITADKQKNGIKANFKIRHNIEDGSVQLADHYQQNTPIGDG PVLLPDNHYLSTQSALSKDPNEKRDHMVLLEFVTAAGITHGMDELYKHHHHHH* >P1;1qyo structureX:1qyo.pdb:1:A:236:A:::: --------------------------SKGEELFTGVVPILVELDGDVNGHKFSVSGEGEGDATYGKLTLKFI CTTGKLPVPWPTLVTTLGGGVQCFSRYPDHMKQHDFFKSAMPEGYVQERTISFKDDGNYKTRAEVKFEGDTL VNRIELKGIDFKEDGNILGHKLEYNYNSHNVYITADKQKNGIKANFKIRHNIEDGSVQLADHYQQNTPIGDG PVLLPDNHYLSTQSALSKDPNEKRDHMVLLEFVTAAGITHGMDELY-------* >P1;1RKK_1 structureX:1RKK_1.pdb:1:A:18:A:::: -RRWCFRVCYRGFCYRKCR----------------------------------------------------- ------------------------------------------------------------------------ ------------------------------------------------------------------------ -----------------------------------------------------* >P1;1HST-T22 structureX:1HST-T22.pdb:40:A:49:A:::: ----------------KSRGGSSRQS---------------------------------------------- ------------------------------------------------------------------------ ------------------------------------------------------------------------ -----------------------------------------------------* >P1;2abx structureX:2abx: 1 :A:74 :B:bungarotoxin:bungarus multicinctus:2.5:-1.00 IVCHTTATIPSSAVTCPPGENLCYRKMWCDAFCSSRGKVVELGCAATCPSKKPYEEVTCCSTDKCNHPPKRQPG/ IVCHTTATIPSSAVTCPPGENLCYRKMWCDAFCSSRGKVVELGCAATCPSKKPYEEVTCCSTDKCNHPPKRQPG* >P1;1hc9 sequence:1hc9: 1 :A:148:B:undefined:undefined:-1.00:-1.00 IVCHTTATSPISAVTCPPGENLCYRKMWCDVFCSSRGKVVELGCAATCPSKKPYEEVTCCSTDKCNPHPKQRPG/ IVCHTTATSPISAVTCPPGENLCYRKMWCDAFCSSRGKVVELGCAATCPSKKPYEEVTCCSTDKCNPHPKQRPG*
Fastq https://en.wikipedia.org/wiki/FASTQ_format Line 1 begins with a '@' character and is followed by a sequence identifier and an optional description. This line format depends on the source (like a FASTA title line). Line 2 is the raw sequence letters. Line 3 begins with a '+' character and is optionally followed by the same sequence identifier (and any description) again. Line 4 encodes the quality values for the sequence in Line 2, and must contain the same number of symbols as letters in the sequence. This symbols are numbers, not in base 10 but in a different base depending on the sequencing system (eg: base 41 for Illumina, base 93 for PacBio, etc) @SEQ_ID GATTTGGGGTTCAAAGCAGTATCGATCAAATAGTAAATCCATTTGTTCAACTCACAGTTT + !''*((((***+))%%%++)(%%%%).1***-+*''))**55CCF>>>>>>CCCCCCC65 @SEQ_ID GGGTGATGGCCGCTGCCGATGGCGTCAAATCCCACC + !"#$%&'()*+,-./0123456789:;<=>?@ABCD
GenBank Flat File Format (gbff) AKA gb, gp, gbk https://www.ncbi.nlm.nih.gov/Sitemap/samplerecord.htm LOCUS HUMCYPB 1868 bp mRNA linear PRI 27-APR-1993 DEFINITION Human cytochrome P-450 1 mRNA, complete cds, clone Hp1-2. ACCESSION M17398 J03472 SOURCE Homo sapiens (human) ORGANISM Homo sapiens Eukaryota; Metazoa; Chordata; Craniata; Vertebrata; Euteleostomi; Mammalia; Eutheria; Euarchontoglires; Primates; Haplorrhini; Catarrhini; Hominidae; Homo. REFERENCE 1 (bases 1 to 1868) AUTHORS Okino,S.T., Quattrochi,L.C., Pendurthi,U.R., McBride,O.W. and Tukey,R.H. TITLE Characterization of multiple human cytochrome P-450 1 cDNAs. The chromosomal localization of the gene and evidence for alternate RNA splicing JOURNAL J. Biol. Chem. 262 (33), 16072-16079 (1987) FEATURES Location/Qualifiers source 1..1868 /organism="Homo sapiens" /mol_type="mRNA" /db_xref="taxon:9606" CDS 2..1183 /note="cytochrome P-450 1" /codon_start=1 /protein_id="AAA35740.1" /db_xref="GI:181328" /translation="MEPFVVLVLCLSFMLLFSLWRQSCRRRKLPPGPTPLPIIGNMLQ IDVKDICKSFTNFSKVYGPVFTVYFGMNPIVVFHGYEAVKEALIDNGEEFSGRGNSPI SQRITKGLGIISSNGKRWKEIRRFSLTNLRNFGMGKRSIEDRVQEEAHCLVEELRKTK ASPCDPTFILGCAPCNVICSVVFQKRFDYKDQNFLTLMKRFNENFRILNSPWIQVCNN FPLLIDCFPGTHNKVLKNVALTRSYIREKVKEHQASLDVNNPRDFMDCFLIKMEQEKD NQKSEFNIENLVGTVADLFVAGTETTSTTLRYGLLLLLKHPEVTAKVQEEIDHVIGRH RSPCMQDRSHMPYTDAVVHEIQRYSDLVPTGVPHAVTTDTKFRNYLIPKSFDNKIMLA A" ORIGIN 337 bp 1 aatggaacct tttgtggtcc tggtgctgtg tctctctttt atgcttctct tttcactctg 61 gagacagagc tgtaggagaa ggaagctccc tcctggcccc actcctcttc ctattattgg 121 aaatatgcta cagatagatg ttaaggacat ctgcaaatct ttcaccaatt tctcaaaagt 181 ctatggtcct gtgttcaccg tgtattttgg catgaatccc atagtggtgt ttcatggata 241 tgaggcagtg aaggaagccc tgattgataa tggagaggag ttttctggaa gaggcaattc 301 cccaatatct caaagaatta ctaaaggact tggaatcatt tccagcaatg gaaagagatg 361 gaaggagatc cggcgtttct ccctcacaaa cttgcggaat tttgggatgg ggaagaggag 421 cattgaggac cgtgttcaag aggaagctca ctgccttgtg gaggagttga gaaaaaccaa 481 ggcttcaccc tgtgatccca ctttcatcct gggctgtgct ccctgcaatg tgatctgctc 541 cgttgttttc cagaaacgat ttgattataa agatcagaat tttctcaccc tgat...... // Identification codes accession organism division feature definition: http://www.insdc.org/files/feature_table.html Source and References Both Nucleotides (gb) or Peptides (gp) Do not confuse the format with the Database Can contain annotated information over a sequence (genes, CDS, regions, etc.) journal Identifiers CDS coordinates protein name Feature Table type of molecule protein sequence DNA sequence
ID M17398; SV 1; linear; mRNA; STD; HUM; 1868 BP. XX AC M17398; J03472; XX DT 20-FEB-1989 (Rel. 18, Created) DT 02-JUL-1999 (Rel. 60, Last updated, Version 6) XX DE Human cytochrome P-450 1 mRNA, complete cds, clone Hp1-2. XX KW alternative splicing; cytochrome P450. XX OS Homo sapiens (human) OC Eukaryota; Metazoa; Chordata; Craniata; Vertebrata; Euteleostomi; Mammalia; OC Eutheria; Euarchontoglires; Primates; Haplorrhini; Catarrhini; Hominidae; OC Homo. XX RN [1] RP 1-1868 RX PUBMED; 3500169. RA Okino S.T., Quattrochi L.C., Pendurthi U.R., McBride O.W., Tukey R.H.; RT "Characterization of multiple human cytochrome P-450 1 cDNAs. The RT chromosomal localization of the gene and evidence for alternate RNA RT splicing"; RL J Biol Chem 262(33):16072-16079(1987). XX DR MD5; 902513e27746967a5785df366127e9d6. DR Ensembl-Gn; ENSG00000138115; homo_sapiens. DR Ensembl-Tr; ENST00000371270; homo_sapiens. DR Ensembl-Tr; ENST00000535898; homo_sapiens. DR EuropePMC; PMC5719403; 29215029. XX CC Draft entry and computer-readable sequence for [1] kindly provided CC by R.Tukey, 02-OCT-1987. CC The two mRNAs encoding citochrome P-450 1 were derived from the CC same gene by alternative splicing. A 39 bp segment of intron 7 CC appears in the sequence below at positions 1151-1189. The other CC mRNA is to be found under accession M17397. CC A polyadenylation signal is located at positions 1840-1845. XX FH Key Location/Qualifiers FH FT source 1..1868 FT /organism="Homo sapiens" FT /mol_type="mRNA" FT /db_xref="taxon:9606" FT CDS 2..1183 FT /codon_start=1 FT /note="cytochrome P-450 1" FT /db_xref="GOA:P10632" FT /db_xref="H-InvDB:HIT000194540.14" FT /db_xref="HGNC:HGNC:2622" FT /db_xref="InterPro:IPR001128" FT /db_xref="InterPro:IPR002401" FT /db_xref="InterPro:IPR017972" FT /db_xref="InterPro:IPR036396" FT /db_xref="PDB:1PQ2" FT /db_xref="PDB:2NNH" FT /db_xref="PDB:2NNI" FT /db_xref="PDB:2NNJ" FT /db_xref="PDB:2VN0" FT /db_xref="UniProtKB/Swiss-Prot:P10632" FT /protein_id="AAA35740.1" FT /translation="MEPFVVLVLCLSFMLLFSLWRQSCRRRKLPPGPTPLPIIGNMLQI FT DVKDICKSFTNFSKVYGPVFTVYFGMNPIVVFHGYEAVKEALIDNGEEFSGRGNSPISQ FT RITKGLGIISSNGKRWKEIRRFSLTNLRNFGMGKRSIEDRVQEEAHCLVEELRKTKASP FT CDPTFILGCAPCNVICSVVFQKRFDYKDQNFLTLMKRFNENFRILNSPWIQVCNNFPLL FT IDCFPGTHNKVLKNVALTRSYIREKVKEHQASLDVNNPRDFMDCFLIKMEQEKDNQKSE FT FNIENLVGTVADLFVAGTETTSTTLRYGLLLLLKHPEVTAKVQEEIDHVIGRHRSPCMQ FT DRSHMPYTDAVVHEIQRYSDLVPTGVPHAVTTDTKFRNYLIPKSFDNKIMLAA" XX SQ Sequence 1868 BP; 548 A; 413 C; 369 G; 538 T; 0 other; aatggaacct tttgtggtcc tggtgctgtg tctctctttt atgcttctct tttcactctg 60 gagacagagc tgtaggagaa ggaagctccc tcctggcccc actcctcttc ctattattgg 120 aaatatgcta cagatagatg ttaaggacat ctgcaaatct ttcaccaatt tctcaaaagt 180 ctatggtcct gtgttcaccg tgtattttgg catgaatccc atagtggtgt ttcatggata 240 tgaggcagtg aaggaagccc tgattgataa tggagaggag ttttctggaa gaggcaattc 300 cccaatatct caaagaatta ctaaaggact tggaatcatt tccagcaatg gaaagagatg 360 gaaggagatc cggcgtttct ccctcacaaa cttgcggaat tttgggatgg ggaagaggag 420 cattgaggac cgtgttcaag aggaagctca ctgccttgtg gaggagttga gaaaaaccaa 480 ggcttcaccc tgtgatccca ctttcatcct gggctgtgct ccctgcaatg tgatctgctc 540 cgttgttttc cagaaacgat ttgattataa agatcagaat tttctcaccc tgatgaaaag 600 attcaatgaa aacttcagga ttctgaactc cccatggatc caggtctgca ataatttccc 660 tctactcatt gattgtttcc caggaactca caacaaagtg cttaaaaatg ttgctcttac 720 acgaagttac attagggaga aagtaaaaga acaccaagca tcactggatg ttaacaatcc 780 tcgggacttt atggattgct tcctgatcaa aatggagcag gaaaaggaca accaaaagtc 840 agaattcaat attgaaaact tggttggcac tgtagctgat ctatttgttg ctggaacaga 900 gacaacaagc accactctga gatatggact cctgctcctg ctgaagcacc cagaggtcac 960 agctaaagtc caggaagaga ttgatcatgt aattggcaga cacaggagcc cctgcatgca 1020 ggataggagc cacatgcctt acactgatgc tgtagtgcac gagatccaga gatacagtga 1080 ccttgtcccc accggtgtgc cccatgcagt gaccactgat actaagttca gaaactacct 1140 catccccaag agctttgata acaagataat gctggctgca taaaactagg gcacaaccat 1200 aatggcatta ctgacttccg tgctacatga tgacaaagaa tttcctaatc caaatatctt 1260 tgaccctggc cactttctag ataagaatgg caactttaag aaaagtgact acttcatgcc 1320 tttctcagca ggaaaacgaa tttgtgcagg agaaggactt gcccgcatgg agctattttt 1380 atttctaacc acaattttac agaactttaa cctgaaatct gttgatgatt taaagaacct 1440 caatactact gcagttacca aagggattgt ttctctgcca ccctcatacc agatctgctt 1500 catccctgtc tgaagaatgc tagcccatct ggctgctgat ctgctatcac ctgcaactct 1560 ttttttatca aggacattcc cactattatg tcttctctga cctctcatca aatcttccca 1620 ttcactcaat atcccataag catccaaact ccattaagga gagttgttca ggtcactgca 1680 caaatatatc tgcaattatt catactctgt aacacttgta ttaattgctg catatgctaa 1740 tacttttcta atgctgactt tttaatatgt tatcactgta aaacacagaa aagtgattaa 1800 tgaatgataa tttagtccat ttcttttgtg aatgtgctaa ataaaaagtg ttattaattg 1860 ctggttca 1868 // EMBL and UNIPOT txt data file ENA: https://ena- docs.readthedocs.io/en/latest/submit/fil eprep/flat-file-example.html UNIPROT: https://web.expasy.org/docs/userma n.html feature definition: http://www.insdc.org/files/feature_t able.html Very similar to gbff, with the same information. Do not confuse the format with the Database or the organization. Same information as Continues next column:
GFF3 https://www.ensembl.org/info/website/upload/gff3.html Several DBs present annotation data in sequences in this format which has become a standard Can include the sequence, but it usually does not ##sequence-region M17398.1 1 1868 ##species https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=9606 M17398.1 Genbank region M17398.1 Genbank CDS 1 2 1868 1183 . . + + . 0 ID=M17398.1:1..1868;Dbxref=taxon:9606;gbkey=Src;mol_type=mRNA ID=cds-AAA35740.1;Dbxref=NCBI_GP:AAA35740.1;Name=AAA35740.1;gbkey=CDS;product=AAA35740.1;protein_id
######################################## # Program: water # Rundate: Wed 14 Sep 2022 13:36:24 # Commandline: water # -auto # -stdout # -asequence emboss_water-I20220914-133154-0897-82573330-p1m.asequence # -bsequence emboss_water-I20220914-133154-0897-82573330-p1m.bsequence # -datafile EBLOSUM62 # -gapopen 10.0 # -gapextend 0.5 # -aformat3 pair # -sprotein1 # -sprotein2 # Align_format: pair # Report_file: stdout ######################################## Program parameters Pair format pairwise aligned sequences. #======================================= # # Aligned_sequences: 2 # 1: EMBOSS_001 # 2: EMBOSS_002 # Matrix: EBLOSUM62 # Gap_penalty: 10.0 # Extend_penalty: 0.5 # # Length: 141 # Identity: 34/141 (24.1%) # Similarity: 54/141 (38.3%) # Gaps: 20/141 (14.2%) # Score: 44.0 # # #======================================= sequences details Alignment and EMBOSS_001 7 VIQIGRQRFVGGLFWQSLSRRNELRAEAVELAKKL---KFDLMVLRIDRG 53 |..||.:::...|.|.| |:..|...:|:..:..| |...::.|.... EMBOSS_002 8 VAVIGSKQYAVNLLWGS-SQDTETTNQALNKSLTLMSSKLYSVIGRFQGE 56 "|": Identical ".": Similar ":": Very similar " ": different not similar EMBOSS_001 54 VAAAGYANTRDGFAPGHLSLGAMVSRAIALEGAFYNGRRQPAPN--WLGA 101 ..|.|..|. ||......:..||..:|:.:.| ..||.| ||.. EMBOSS_002 57 QFAVGDKNI------GHKRGQVTLLSAIDFDGSSFCG-LFPADNELWLVI 99 EMBOSS_001 102 FALPDGRWAYFAVRDHAFMPNGDWVGSREEALERLHTDYAW 142 ....|| ..:| |.:|....| :::...:.:...|.| EMBOSS_002 100 GVDKDG-MVHF---DKSFHSKDD---AKKFFFDHVAYGYPW 133 #--------------------------------------- #---------------------------------------
RID: J3M9FXS001R Job Title:2QGU_1|Chain A|Probable signal peptide protein|Ralstonia... Program: BLASTP Database: pdb PDB protein database Query #1: 2QGU_1|Chain A|Probable signal peptide protein|Ralstonia solanacearum (267608) Query ID: lcl|Query_117944 Length: 211 blast format Results from blast search. Mulitple pairwaise alignments short description job details and Sequences producing significant alignments: Scientific Common Max Total Query Description Name Name Taxid Score Score cover Value Ident Len Accession Chain A, Periplasmic transport protein [Neisseria gonorrhoeae... Neisseria go... NA 521006 83.2 83.2 71% 2e-19 33.12 183 8DTE_A Two-phospholipid-bound crystal structure of the... Pseudomonas ... NA 208964 65.5 65.5 70% 1e-12 27.81 207 6HSY_A Crystal Structure of Toluene-tolerance protein from Pseudomona... Pseudomonas ... NA 160488 62.4 62.4 72% 2e-11 24.52 223 4FCZ_A of results E Per. Acc. Alignments: >Chain A, Periplasmic transport protein [Neisseria gonorrhoeae NCCP11945] Sequence ID: 8DTE_A Length: 183 Range 1: 29 to 180 Subject sequence data Score:83.2 bits(204), Expect:2e-19, Method:Compositional matrix adjust., Identities:51/154(33%), Positives:82/154(53%), Gaps:5/154(3%) Alignement data Alignment: "X" : AA code indicates Identical "+" : Positive value in the used matrix ( similar) " " : Different not positive Query 52 LRGGNLQKVFQLVDQKIVPRADFKRTTQIAMGRFWSQATPEQQQQIQDGFKSLLIRTYAG 111 L+ G+ + VP DF+R T +A+G W A+ Q+Q + F++LLIRTY+G Sbjct 29 LKSGDAASARPKAEAYAVPYFDFQRMTALAVGNPWRTASDAQKQALAKEFQTLLIRTYSG 88 Query 112 ALANVRNQTVAYK--PFRAAADDTDVVVRSTVNNNGE-PVALDYRVEKSPNGWKVYDINI 168 + +N TV K P ++VVR+ V G+ PV +D+ +S ++ Y++ I Sbjct 89 TMLKFKNATVNVKDNPI-VNKGGKEIVVRAEVGIPGQKPVNMDFTTYQSGGKYRTYNVAI 147 Query 169 SGLWLSETYKNQFADVISKRGGVGGLVQFLDERN 202 G L Y+NQF ++I K G+ GL+ L +N Sbjct 148 EGTSLVTVYRNQFGEII-KAKGIDGLIAELKAKN 180 >Two-phospholipid-bound crystal structure of the substrate-binding protein Ttg2D from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1] Sequence ID: 6HSY_A Length: 207 Range 1: 13 to 163 Score:65.5 bits(158), Expect:1e-12, Method:Compositional matrix adjust., Identities:42/151(28%), Positives:80/151(52%), Gaps:2/151(1%) Query 39 VDDVLATIKGDPDLRGGNLQKVFQLVDQKIVPRADFKRTTQIAMG-RFWSQATPEQQQQI 97 VD++L+ IK + + QK++ +D+ + P D + + M ++ QA+PEQ ++ Sbjct 13 VDELLSDIKANKAAYKADPQKLYATLDRILGPVVDAEGIAKSVMTVKYSRQASPEQIKRF 72 Query 98 QDGFKSLLIRTYAGALANVRNQTVAYKPFRA-AADDTDVVVRSTVNNNGEPVALDYRVEK 156 ++ FK+ L++ Y AL NQ + P A +DD V ++ G + Y + Sbjct 73 EEVFKNSLMQFYGNALLEYDNQDIRVLPSSAKPSDDRASVNMEIRDSKGTVYPVSYTMTN 132
blast 6 format (AKA Hit table text) Simplified results from blast search. data results from pairwaise alignments # blastp # Iteration: 0 # Query: 2QGU_1|Chain A|Probable signal peptide protein|Ralstonia solanacearum (267608) # RID: J3M9FXS001R # Database: pdb # Fields: query acc.ver, subject acc.ver, % identity, alignment length, mismatches, gap opens, q. start, q. end, s. start, s. end, evalue, bit score, % positives # 4 hits found 2QGU_1|Chain 2QGU_A 99.524 210 1 0 2 2QGU_1|Chain 8DTE_A 33.117 154 98 4 52 2QGU_1|Chain 6HSY_A 27.815 151 107 2 39 2QGU_1|Chain 4FCZ_A 24.516 155 116 1 36 Job description 211 202 187 189 2 29 13 31 211 180 163 185 4.05e-135 1.81e-19 1.10e-12 2.13e-11 378 83.2 65.5 62.4 99.52 53.25 52.98 47.74 Fileds (columns) description
clustalw format Most used format for multiple sequence alignments CLUSTAL O(1.2.4) multiple sequence alignment 6HSY_A ---------------------MAPTPQ--------QVVQGTVDELLSDIKANKAAYKADP 4FCZ_A XISILRRGLLVLLAAFPLLALAVQTPH--------EVVQSTTNELLGDLKANKEQYKSNP 2QGU_A ---XFKKLLHSLVAGLTFVAAVAAVPAHAQEADAQATVKTAVDDVLATIKGDPDLRGGNL 8DTE_A --------------------MAHHHHHHMSPADAVGQIRQNATQVLTIL------KSGDA :: . ::* : .: "*" Identical position ":" Very similar "." Similar " " Different not similar 6HSY_A QKLYATLDRILGPVVDAEGIAKSVMTVKYSRQASPEQIKRFEEVFKNSLMQFYGNALLEY 4FCZ_A NAFYDSLNRILGPVVDADGISRSIXTVKYSRKATPEQXQRFQENFKRSLXQFYGNALLEY 2QGU_A QKVFQLVDQKIVPRADFKRTT-QIAXGRFWSQATPEQQQQIQDGFKSLLIRTYAGALANV 8DTE_A ASARPKAEAYAVPYFDFQRMT-ALAVGNPWRTASDAQKQALAKEFQTLLIRTYSGTMLKF : * * . : : . *: * : : . *: * : *..:: : 6HSY_A DNQDIRVLPSSAKP-SDDRASVNMEIRDSKGTVYPVSYTMTNLAGGWKVRNVIINGINIG 4FCZ_A NNQGITVDPAKAD--DGKRASVGXKVTGNNGAVYPVQYTLENIGGEWKVRNVIVNGINIG 2QGU_A RNQTVAYKPFRAAA-DDTDVVVRSTVN-NNGEPVALDYRVEKSPNGWKVYDINISGLWLS 8DTE_A KNATVNVKDNPIVNKGGKEIVVRAEVGIPGQKPVNMDFTTYQSGGKYRTYNVAIEGTSLV * : .. * : :.: : . ::. :: :.* : 6HSY_A KLFRDQFADTMQKNRNDLEKTIAGWGEVVAKAKETAKAEEAGAKKLAAALEHHHHHH 4FCZ_A KLFRDQFADAXQRNGNDLDKTIDGWAGEVAKAKQAADNSPEKSVKLEHHHHHH---- 2QGU_A ETYKNQFADVISKRGG--------VGGLVQFLDERNAQLAKGPAK------------ 8DTE_A TVYRNQFGEIIKAKGI---------DGLIAELKAKNGGK------------------ :::**.: . . : .
phylip format multiple sequence alignments Number of seqs Number of positions in the alignment interleaved Max 10 characters as sequence name 4 237 I 2QGU_A ---XFKKLLH SLVAGLTFVA AVAAVPAHAQ EADAQATVKT AVDDVLATIK GDPDLRGGNL QKVFQLVDQK IVPRADFKRT T-QIAXGRFW SQATPEQQQQ IQDGFKSLLI RTYAGALANV RNQTVAYKPF RAAA-DDTDV VVRSTVN-NN GEPVALDYRV EKSPNGWKVY DINISGLWLS ETYKNQFADV ISKRGG---- ----VGGLVQ FLDERNAQLA KGPAK----- ------- The sequences may be written in one long stretch, divided into several lines, or in blocks interleaved with blanks (I) 8DTE_A ---------- ---------- MAHHHHHHMS PADAVGQIRQ NATQVLTIL- -----KSGDA ASARPKAEAY AVPYFDFQRM T-ALAVGNPW RTASDAQKQA LAKEFQTLLI RTYSGTMLKF KNATVNVKDN PIVNKGGKEI VVRAEVGIPG QKPVNMDFTT YQSGGKYRTY NVAIEGTSLV TVYRNQFGEI IKAKGI---- -----DGLIA ELKAKNGGK- ---------- ------- 6HSY_A ---------- ---------- -MAPTPQ--- -----QVVQG TVDELLSDIK ANKAAYKADP QKLYATLDRI LGPVVDAEGI AKSVMTVKYS RQASPEQIKR FEEVFKNSLM QFYGNALLEY DNQDIRVLPS SAKP-SDDRA SVNMEIRDSK GTVYPVSYTM TNLAGGWKVR NVIINGINIG KLFRDQFADT MQKNRNDLEK TIAGWGEVVA KAKETAKAEE AGAKKLAAAL EHHHHHH 4FCZ_A XISILRRGLL VLLAAFPLLA LAVQTPH--- -----EVVQS TTNELLGDLK ANKEQYKSNP NAFYDSLNRI LGPVVDADGI SRSIXTVKYS RKATPEQXQR FQENFKRSLX QFYGNALLEY NNQGITVDPA KAD--DGKRA SVGXKVTGNN GAVYPVQYTL ENIGGEWKVR NVIVNGINIG KLFRDQFADA XQRNGNDLDK TIDGWAGEVA KAKQAADNSP EKSVKLEHHH HHH----
nexus format multiple sequence alignments with extra parameters Multiple extra fields can be included designed to be read by specific software (eg: MrBayes, Splitstree, etc) #NEXUS BEGIN DATA; DIMENSIONS NTAX=4 NCHAR=237; FORMAT DATATYPE=PROTEIN INTERLEAVE MISSING=-; [Name: 2QGU_A Len: 237 Check: 0] [Name: 8DTE_A Len: 237 Check: 0] [Name: 6HSY_A Len: 237 Check: 0] [Name: 4FCZ_A Len: 237 Check: 0] MATRIX 2QGU_A ---XFKKLLHSLVAGLTFVA AVAAVPAHAQEADAQATVKT AVDDVLATIKGDPDLRGGNL QKVFQLVDQKIVPRADFKRT T-QIAXGRFWSQATPEQQQQ 8DTE_A -------------------- MAHHHHHHMSPADAVGQIRQ NATQVLTIL------KSGDA ASARPKAEAYAVPYFDFQRM T-ALAVGNPWRTASDAQKQA 6HSY_A -------------------- -MAPTPQ--------QVVQG TVDELLSDIKANKAAYKADP QKLYATLDRILGPVVDAEGI AKSVMTVKYSRQASPEQIKR 4FCZ_A XISILRRGLLVLLAAFPLLA LAVQTPH--------EVVQS TTNELLGDLKANKEQYKSNP NAFYDSLNRILGPVVDADGI SRSIXTVKYSRKATPEQXQR 2QGU_A IQDGFKSLLIRTYAGALANV RNQTVAYKPFRAAA-DDTDV VVRSTVN-NNGEPVALDYRV EKSPNGWKVYDINISGLWLS ETYKNQFADVISKRGG---- 8DTE_A LAKEFQTLLIRTYSGTMLKF KNATVNVKDNPIVNKGGKEI VVRAEVGIPGQKPVNMDFTT YQSGGKYRTYNVAIEGTSLV TVYRNQFGEIIKAKGI---- 6HSY_A FEEVFKNSLMQFYGNALLEY DNQDIRVLPSSAKP-SDDRA SVNMEIRDSKGTVYPVSYTM TNLAGGWKVRNVIINGINIG KLFRDQFADTMQKNRNDLEK 4FCZ_A FQENFKRSLXQFYGNALLEY NNQGITVDPAKAD--DGKRA SVGXKVTGNNGAVYPVQYTL ENIGGEWKVRNVIVNGINIG KLFRDQFADAXQRNGNDLDK 2QGU_A ----VGGLVQFLDERNAQLA KGPAK------------ 8DTE_A -----DGLIAELKAKNGGK- ----------------- 6HSY_A TIAGWGEVVAKAKETAKAEE AGAKKLAAALEHHHHHH 4FCZ_A TIDGWAGEVAKAKQAADNSP EKSVKLEHHHHHH---- ; END;
json format https://en.wikipedia.org/wiki/JSON Not a bioinformatics specific format Most used format to exchange data in web services Heavily used for bioinformatics (it is similar to a python dictionary or a perl hash) { "firstName": "John", "lastName": "Smith", "isAlive": true, "age": 27, "address": { "streetAddress": "21 2nd Street", "city": "New York", "state": "NY", "postalCode": "10021-3100" }, "phoneNumbers": [ { "type": "home", "number": "212 555-1234" }, { "type": "office", "number": "646 555-4567" } ], "children": [ "Catherine", "Thomas", "Trevor" ], "spouse": null } "key" : "value" (value is a string or number) "key" : { (value is a dictionary) "key" : [ (value is a list)






















