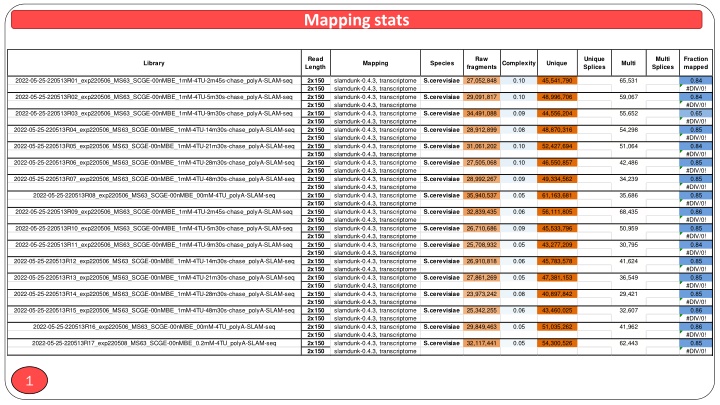
Mapping Stats and Complexity Analysis of SLAM-seq Data
In this dataset, raw input reads and unique splice variants of S. cerevisiae samples were analyzed through SLAM-seq experiments, showcasing different time points during a "chase" period. The analysis includes mapped fraction percentages, library complexity assessments, and both unique and multi-mapping fragments. Variants in Cln3 and Whi5 are also explored, providing valuable insights into the gene expression dynamics under experimental conditions. Additionally, a visual representation of the mapping statistics and key findings is available for a comprehensive understanding of the data.
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
You are allowed to download the files provided on this website for personal or commercial use, subject to the condition that they are used lawfully. All files are the property of their respective owners.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.
E N D
Presentation Transcript
Mapping stats Read Length Raw Unique Splices Multi Splices Fraction mapped Library Mapping Species Complexity Unique Multi fragments 2022-05-25-220513R01_exp220506_MS63_SCGE-00nMBE_1mM-4TU-2m45s-chase_polyA-SLAM-seq slamdunk-0.4.3, transcriptome slamdunk-0.4.3, transcriptome slamdunk-0.4.3, transcriptome slamdunk-0.4.3, transcriptome slamdunk-0.4.3, transcriptome slamdunk-0.4.3, transcriptome slamdunk-0.4.3, transcriptome slamdunk-0.4.3, transcriptome slamdunk-0.4.3, transcriptome slamdunk-0.4.3, transcriptome slamdunk-0.4.3, transcriptome slamdunk-0.4.3, transcriptome slamdunk-0.4.3, transcriptome slamdunk-0.4.3, transcriptome slamdunk-0.4.3, transcriptome slamdunk-0.4.3, transcriptome slamdunk-0.4.3, transcriptome slamdunk-0.4.3, transcriptome slamdunk-0.4.3, transcriptome slamdunk-0.4.3, transcriptome slamdunk-0.4.3, transcriptome slamdunk-0.4.3, transcriptome slamdunk-0.4.3, transcriptome slamdunk-0.4.3, transcriptome slamdunk-0.4.3, transcriptome slamdunk-0.4.3, transcriptome slamdunk-0.4.3, transcriptome slamdunk-0.4.3, transcriptome slamdunk-0.4.3, transcriptome slamdunk-0.4.3, transcriptome slamdunk-0.4.3, transcriptome slamdunk-0.4.3, transcriptome slamdunk-0.4.3, transcriptome slamdunk-0.4.3, transcriptome 27,052,848 0.10 45,541,790 65,531 0.84 #DIV/0! 0.84 #DIV/0! 0.65 #DIV/0! 0.85 #DIV/0! 0.84 #DIV/0! 0.85 #DIV/0! 0.85 #DIV/0! 0.85 #DIV/0! 0.86 #DIV/0! 0.85 #DIV/0! 0.84 #DIV/0! 0.85 #DIV/0! 0.85 #DIV/0! 0.85 #DIV/0! 0.86 #DIV/0! 0.86 #DIV/0! 0.85 #DIV/0! 2x150 2x150 2x150 2x150 2x150 2x150 2x150 2x150 2x150 2x150 2x150 2x150 2x150 2x150 2x150 2x150 2x150 2x150 2x150 2x150 2x150 2x150 2x150 2x150 2x150 2x150 2x150 2x150 2x150 2x150 2x150 2x150 2x150 2x150 S.cerevisiae 2022-05-25-220513R02_exp220506_MS63_SCGE-00nMBE_1mM-4TU-5m30s-chase_polyA-SLAM-seq 29,091,817 0.10 48,996,706 59,067 S.cerevisiae 2022-05-25-220513R03_exp220506_MS63_SCGE-00nMBE_1mM-4TU-9m30s-chase_polyA-SLAM-seq 34,491,088 0.09 44,556,204 55,652 S.cerevisiae 2022-05-25-220513R04_exp220506_MS63_SCGE-00nMBE_1mM-4TU-14m30s-chase_polyA-SLAM-seq 28,912,899 0.08 48,870,316 54,298 S.cerevisiae 2022-05-25-220513R05_exp220506_MS63_SCGE-00nMBE_1mM-4TU-21m30s-chase_polyA-SLAM-seq 31,061,202 0.10 52,427,694 51,064 S.cerevisiae Cln3 Whi5 switched!!! 2022-05-25-220513R08_exp220506_MS63_SCGE-00nMBE_00mM-4TU_polyA-SLAM-seq 2022-05-25-220513R06_exp220506_MS63_SCGE-00nMBE_1mM-4TU-28m30s-chase_polyA-SLAM-seq 27,505,068 0.10 46,550,857 42,486 S.cerevisiae 2022-05-25-220513R07_exp220506_MS63_SCGE-00nMBE_1mM-4TU-48m30s-chase_polyA-SLAM-seq 28,992,267 0.09 49,334,562 34,239 S.cerevisiae 35,940,537 0.05 61,163,681 35,686 S.cerevisiae 2022-05-25-220513R09_exp220506_MS63_SCGE-00nMBE_1mM-4TU-2m45s-chase_polyA-SLAM-seq 32,839,435 0.06 56,111,805 68,435 S.cerevisiae 2022-05-25-220513R10_exp220506_MS63_SCGE-00nMBE_1mM-4TU-5m30s-chase_polyA-SLAM-seq 26,710,686 0.09 45,533,796 50,959 S.cerevisiae 2022-05-25-220513R11_exp220506_MS63_SCGE-00nMBE_1mM-4TU-9m30s-chase_polyA-SLAM-seq 25,708,932 0.05 43,277,209 30,795 S.cerevisiae 2022-05-25-220513R12_exp220506_MS63_SCGE-00nMBE_1mM-4TU-14m30s-chase_polyA-SLAM-seq 26,910,818 0.06 45,783,578 41,624 S.cerevisiae 2022-05-25-220513R13_exp220506_MS63_SCGE-00nMBE_1mM-4TU-21m30s-chase_polyA-SLAM-seq 27,861,269 0.05 47,381,153 36,549 S.cerevisiae 2022-05-25-220513R14_exp220506_MS63_SCGE-00nMBE_1mM-4TU-28m30s-chase_polyA-SLAM-seq 23,973,242 0.08 40,897,842 29,421 S.cerevisiae 2022-05-25-220513R15_exp220506_MS63_SCGE-00nMBE_1mM-4TU-48m30s-chase_polyA-SLAM-seq 25,342,255 0.06 43,460,025 32,607 S.cerevisiae 2022-05-25-220513R16_exp220506_MS63_SCGE-00nMBE_00mM-4TU_polyA-SLAM-seq 29,849,463 0.05 51,035,262 41,962 S.cerevisiae 2022-05-25-220513R17_exp220508_MS63_SCGE-00nMBE_0.2mM-4TU_polyA-SLAM-seq 32,117,441 0.05 54,300,526 62,443 S.cerevisiae 1
Cln3 Whi5 switched!!! 2
Cln3 Whi5 switched!!! 3
Cln3 Whi5 switched!!! 4
Cln3 Whi5 switched!!! 5