Understanding In Vitro Transcription and Probe Production
In vitro transcription involves utilizing T7 or T3 RNA polymerase to produce multiple single-stranded RNA copies of a gene of interest. These transcripts can be used to create probes for detecting specific RNA sequences. The process often involves labeling with DIG or radioisotopes like 33P or 32P, which are detected using various methods. Hybridization with labeled probes allows for the detection of gene expression levels. More complex methods involve using different labels and fluorophores. This technique has applications in research, diagnostics, and molecular biology.
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author. Download presentation by click this link. If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
E N D
Presentation Transcript
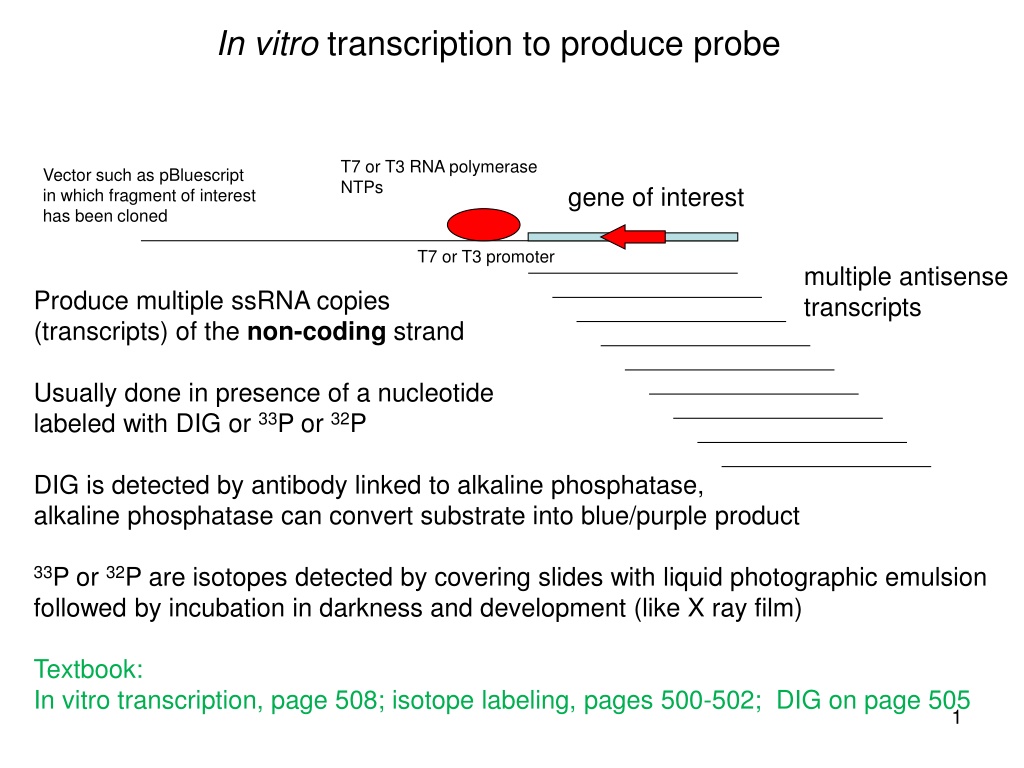
In vitro transcription to produce probe (a) (b) (c) (e) (d) (i) (h) (j) (f) (k) T7 or T3 RNA polymerase NTPs Vector such as pBluescript in which fragment of interest has been cloned gene of interest T7 or T3 promoter multiple antisense transcripts Produce multiple ssRNA copies (transcripts) of the non-coding strand (o) (l) (n) (m) (p) Usually done in presence of a nucleotide labeled with DIG or 33P or 32P DIG is detected by antibody linked to alkaline phosphatase, alkaline phosphatase can convert substrate into blue/purple product (q) (r) 33P or 32P are isotopes detected by covering slides with liquid photographic emulsion followed by incubation in darkness and development (like X ray film) Textbook: In vitro transcription, page 508; isotope labeling, pages 500-502; DIG on page 505 1
Principle: hybridization with labeled probe (either DNA or RNA) which is complementary to cellular RNA to detect internal expression 2
EXAMPLE: http://people.biology.ucsd.edu/davek/intro.html 3
Can do more complicated ones using different labels and varied fluorophores: DIG label Fluor label DNP label Antibodies to each label, conjugated with unique fluorophore 4 https://insitutech.wordpress.com/2015/12/17/in-situ-hybridization/
The type of image that can be generated The labels each fluoresce at different wavelengths to produce distinct colours And the regions of overlap show additional easily distinguished colours: Blue + Red = violet Green + Red = yellow 5
major approach 1: whole-mount in situ hybridization Whole or intact structures are hybridized with probe and thereafter mounted on microscope slides for assessment advantages: gives an immediate overview of expression relatively fast and simple disadvantage: low sensitivity and resolution (penetration of tissues) 6
whole-mount in situ hybridization, example from Drosophila embryos In situ hybridization of wild type Drosophila embryos at different developmental stages for the RNA from a gene called hunchback. From Wikipedia 7
Example: expression of a transcription factor detected by whole-mount expression in leaf primordia (e) (d) (f) (c) (g) (b) (a) (h) (i) (j) (k) (l) (m) (n) (r) (p) (q) (s) * (o) 8 Wenzel et al., Plant Journal 2007 49: 387-