Understanding Normal Reference Range and Descriptive Statistics in Medicine
The concept of normal reference range in medicine, determined by collecting data from a population, helps predict intervals where values are expected to fall. Descriptive statistics like mean, variance, skewness, and kurtosis provide insights into data distributions. Methods for calculating reference ranges include sample mean, standard deviations, and transformations. Raw data examples such as creatinine levels in healthy males showcase statistical analysis methods.
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author. Download presentation by click this link. If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
E N D
Presentation Transcript
Normal Reference Range in Medicine and Skewed Distributions Richard B. Goldstein, Ph.D. Professor of Mathematics (ret.) Providence College Courtesy Professor of Mathematics at FGCU
Normal Reference Range : the set of values in which 95% of the normal population falls within (that is, a 95% prediction interval). It is determined by collecting data from typically over one hundred tests from normal patients.
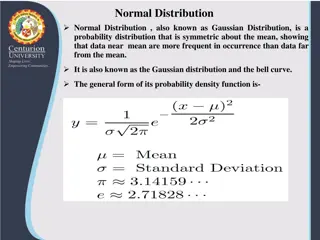
Descriptive Statistics Descriptive Statistics n = x i ( ) = = x Est : sample mean i 1 n ( ) 2 x x ( ) i = = 2 2 s Est : sample var iance n 1 ( ) k x x i = m k n m ( ) = = 3 b Est : skewness 1 1 / 3 2 2 m m ( ) kurtosis : 2 = = b Est 4 2 2 2 m
Common Methods used to calculate the Common Methods used to calculate the Normal Reference Range Normal Reference Range Sample mean 2 standard deviations Percentiles of Raw Data Using general power transformations to create a Normal/Gaussian Distribution Curve fitting of data in histogram format by various skewed distributions Dissecting distributions that contain mixed normal and abnormal populations
Near Gaussian Shaped Data Near Gaussian Shaped Data
Some Raw Data Creatinine (mg/dl) for 100 healthy males # Creatinine # Creatinine # Creatinine # Creatinine # Creatinine 1 0.90 21 0.97 41 1.01 61 1.04 81 1.09 2 0.91 22 0.98 42 1.01 62 1.05 82 1.09 3 0.91 23 0.98 43 1.01 63 1.05 83 1.10 4 0.92 24 0.98 44 1.01 64 1.05 84 1.10 5 0.93 25 0.98 45 1.02 65 1.05 85 1.10 6 0.93 26 0.98 46 1.02 66 1.05 86 1.10 7 0.94 27 0.99 47 1.02 67 1.06 87 1.11 8 0.94 28 0.99 48 1.02 68 1.06 88 1.11 9 0.94 29 0.99 49 1.02 69 1.06 89 1.11 10 0.95 30 0.99 50 1.03 70 1.06 90 1.12 11 0.95 31 0.99 51 1.03 71 1.07 91 1.12 12 0.96 32 1.00 52 1.03 72 1.07 92 1.13 13 0.96 33 1.00 53 1.03 73 1.07 93 1.13 14 0.96 34 1.00 54 1.03 74 1.07 94 1.13 15 0.96 35 1.00 55 1.03 75 1.08 95 1.14 16 0.97 36 1.00 56 1.03 76 1.08 96 1.15 17 0.97 37 1.00 57 1.04 77 1.08 97 1.15 18 0.97 38 1.00 58 1.04 78 1.08 98 1.16 19 0.97 39 1.00 59 1.04 79 1.09 99 1.17 20 0.97 40 1.01 60 1.04 80 1.09 100 1.19
Using ? 1.96? on Raw Data The n sample values are used to find the sample statistics: 0306 . 1 x = = Creatinine 20 s 0644 . 0 18 16 also note 14 12 10 = = 8 skewness b = 2370 . 0 6 1 4 = 5032 . 0 kurtosis b 2 0 2 0.9 0.93 0.96 0.99 1.02 1.05 1.08 1.11 1.14 1.17 1.2 0306 . 1 : s 96 . 1 0644 . 0 ( 96 . 1 x ) Normal Range : . 0 904 to . 1 157
Using Percentiles on Raw Data x x x The n sample values are sorted: n ) 1 ( ) 2 ( ( ) Method I (used by Excel and Quattro Pro for example) ) ( = where x by given p r k k ( ) 1 = + r n 5 . 2 1 100 = = = = + ) 1 = n p p x where r 100 , 5 . 2 1 100 ( . 3 475 r 5 . 2 ( ) 100 97 5 . = = = = + ) 1 = n p p x where r 100 , 97 5 . 1 100 ( 97 525 . r 97 5 . ( ) 100
Method II (used by SPSS and known as Tukeys Hinges) ( ) = = + p given by x where r k n with the following rules 1 : k r ( ) [a] [b] [c] if k(n + 1) < 1 then use r = 1 if k(n + 1) > n then use r = n otherwise interpolate using r = k(n + 1) Using this method the normal range is 0.910 to 1.165 The National Committee for Clinical Laboratory Standards accepted the use of percentiles as an optional method for determining a normal range which was proposed by L. Herrera.
ERRORS IN CLINICAL LABORATORIES Variations between laboratories Variation between analysts (same laboratory site) Day-to-day variations (same analyst, laboratory, and sample) Variations between replicates (same analysis) 0.5 SD 2.0 SD 1.0 SD 20 20 20 15 15 15 Frequency Frequency Frequency 10 10 10 5 5 5 0 0 0 0.75 0.80 0.85 0.90 0.95 1.00 1.05 1.10 1.15 1.20 1.25 0.75 0.80 0.85 0.90 0.95 Creatinine mg/dl 1.00 1.05 1.10 1.15 1.20 1.25 0.75 0.80 0.85 0.90 0.95 Creatinine mg/dl 1.00 1.05 1.10 1.15 1.20 1.25 Creatinine mg/dl
CONVOLUTION (sum of random variables) Z = X + Y X has probability density function f(x) Y has probability density function g(x) Z has probability density function h(x) = h t f x g t x dx ( ) ( ) ( ) Example: Exponential and Normal Distributions 2 x 2 e 2 = = x f x e for x g x all x ( ) , 0 ( ) 2 2 x 2 t 2 2 e t 2 t + = = t x h t e dx e ( ) 1 2 2
Skewed Distributions for Fitting Data Skewed Distributions for Fitting Data Gram Charlier Series Pearson System of Distributions N.L. Johnson System of Distributions Skew-Normal Distribution
Gram Gram- -Charlier Charlier Series Series ( ) 2 1 x x x + + ) x ( f exp 1 H H 1 2 3 4 2 3 4 2 ! 3 ! 4 2 = = + 3 4 2 with H ) x ( 3 x 3 x and H ) x ( 4 x 6 x 3 Problems: 1. f(x) is negative for part of the interval 2. f(x) is bimodal for high skewness
Karl Pearson System of Distributions Karl Pearson System of Distributions ) x ( + f a x ( ) + = 0 + + 2 ) x ( f b b x ( 1 ) b x ( 2 ) 0 Problems: 1. Difficulty in finding percentiles 2. Some complicated forms
N.L. Johnson System of Probability Distributions N.L. Johnson System of Probability Distributions
Highly Skewed Distribution Highly Skewed Distribution A1c/Glycated Hemoglobin A1c/Glycated Hemoglobin x freq A1c - Histogram of n = 474 in NHANES III Study = 1 5 3.25 3.75 4.25 4.75 5.25 5.75 6.25 6.75 7.25 7.75 8.25 8.75 9.25 9.75 10.25 10.75 11.25 11.75 12.25 12.75 n 474 200 17 110 183 102 23 10 = x . 5 505 ( mean ) 180 160 . 1 = s 061 = st ( . dev .) 140 b . 3 459 skew ( ) 1 = 4 4 2 2 1 2 2 1 3 0 1 1 120 b 16 = 256 . ( kurtosis = ) 100 2 . 1 + p x 645 s . 7 250 80 95 60 40 20 0 3.25 3.75 4.25 4.75 5.25 5.75 6.25 6.75 7.25 7.75 8.25 8.75 9.25 9.75 10.25 10.75 11.25 11.75 12.25 12.75 474 TOTAL
Sum Sq = 175.4 P0.95 = 6.06 First Fit First Fit A1c fit by Johnson SU 200 180 Standard: 4% to 5.6% : Normal 5.7% to 6.4% : Pre-Diabetic 6.5% or more : Diabetic 160 140 120 100 80 60 40 20 0 3.25 3.75 4.25 4.75 5.25 5.75 6.25 6.75 7.25 7.75 8.25 8.75 9.25 9.75 10.2510.7511.2511.7512.2512.75 freq fit
Was this a set of healthy individuals only? Was this a set of healthy individuals only? Can we remove some Can we remove some abnormals abnormals/unhealthy /unhealthy? ? A1c - fit by Dissection by Johnson SU & Normal Distributions 200 180 160 140 120 100 80 60 40 20 0 0 0.5 1 1.5 2 2.5 3 3.5 4 4.5 5 5.5 6 6.5 7 7.5 8 8.5 9 9.5 10 10.5 11 11.5 12 12.5 13 13.5 SU-Normals Gauss-Abnormal
Dissected result using Johnson SU & Gaussian Dissected result using Johnson SU & Gaussian Dissection fTot (x)= w1f1(x) + w2f2(x) Sum of squares now 56.71 Approximately 89% of values are healthy (normal) individuals Approximately 11% of values are from unhealthy (abnormal) individuals The 90th percentile is 5.82 The 95th percentile is 5.98 Standard (from sources): 4% to 5.6% : Normal 5.7% to 6.4% : Pre-Diabetic 6.5% or more : Diabetic
Dissection using two Johnson SU distributions Dissection using two Johnson SU distributions Dissection using two Johnson SU distributions 200 190 180 170 160 150 140 130 120 110 100 90 80 70 60 50 40 30 20 10 0 0 0.5 1 1.5 2 2.5 3 3.5 4 4.5 Normal 5 5.5 6 6.5 7 7.5 8 8.5 9 9.5 10 10.5 11 11.5 12 12.5 13 13.5 Abnormal
Dissected result using 2 Johnson SU Distributions Dissected result using 2 Johnson SU Distributions Sum of squares now 37.73 Approximately 91% of values are healthy individuals Approximately 9% of values are from unhealthy individuals The 90th percentile is 5.82 The 95th percentile is 5.99 Almost identical to using Johnson SU with Gaussian Note crossover at 6.4 where we minimize the sum of the false positives and false negatives
Comparisons for A1c Comparisons for A1c 95th Percentile Method Sum of Sqs Simple Normal +1.645s 7.25 Single Johnson SU fit 175.4 6.06 Dissection Johnson SU & Gauss 56.7 5.98 Dissection 2 Johnson SU Current Standard Prediabetic 37.7 5.99 5.60 Current Standard Diabetic 6.50
Highly Skewed Data Highly Skewed Data with Outliers with Outliers
Box Box- -Cox Transformation Cox Transformation i 1 x = 0 y i x ( ) = + = ln 0 y c i i One way to determine, is to select the value of that maximizes the logarithm of the likelihood function (LLF): 2 n n n n n y y 1 ( ) ( ) x = i = i = i = + = , x ( f ) ln 1 ln where y y i i i 2 n 1 1 1
LDH 119 - no transform 30 25 20 15 10 5 0 300 400 500 600 700 800 900 1000 1100 1200 1300 1400 1500 1600 1700 1800 1900 2000 2100 2200 2300 2400 2500 2600 2700 LDH 119 after transform = -0.9742 25 20 15 10 5 0 1.0229 1.0232 1.0235 1.0238 1.0241 1.0244 1.0247 1.0250 1.0253 1.0256 1.0259 1.0262
OUTLIERS in a Normal OUTLIERS in a Normal Distrubtion Distrubtion Prof. John Tukey s Exploratory Data Analysis First Fence beyond [Q1-1.5*IQR] = -2.698 or [Q3+1.5*IQR] = 2.698 about 0.698% totally use symbol Second Fence beyond [Q1-3*IQR] = -4.721 or [Q3+3*IQR] = 4.721 about 0.00023% totally use symbol For LDH where the 1st fence is 1287.6 and 2nd fence is 1761.2 there are 9 outliers of which 4 are extreme outliers this is too many!
Outliers for a lognormal distribution Outliers for a lognormal distribution 2 ln x / 2 e = ) x ( f x 2 Here Q1 = 0.509, Q3 = 1.963 and the 1st fence is 4.143 and 2ndis 6.329. Using Tukey s fences 7.76% of the values are beyond the 1st fence and 3.26% are beyond 2nd fence. For LDH if the top 2 values (4537 and 66592) are dropped, then the 1st fence is at 2873 and there would be no further outliers to remove. An outlier should be errors in measurement or extremely unlikely events. Conclusion: Don t use Tukey s fences for very skewed data. Suggestion: Transform data closer to Normal and then use Tukey s fences to remove outliers.
LDH - Johnson SU 30 30 P2.5 = 375 25 25 20 20 P97.5 = 1432 15 15 10 10 Note: 4 values not fitted are 2327, 2614, 4537, and 66592 5 5 0 0 250 350 450 550 650 750 850 950 1050 1150 1250 1350 1450 1550.0
Comparisons for LDH Comparisons for LDH Method 95% Reference Interval with all 120 LDH values 95% Reference Interval with 116 LDH values Nonparametric Traditional Normal (376, 2334) (0, 13271) (375, 1370) (194, 1138) Transformed Normal (351, 1730) (366, 1351) Least Sq fit by Johnson SU (305, 1104) (375, 1432) Traditional Normal: LDH Data Source: Reference Intervals: A User s Guide by Horn & Pesce AACC Press 2005
Cholesterol Data from the Framingham (MA) Heart Study # 1 2 3 4 5 6 7 8 9 Chol 167 184 192 198 200 202 210 211 212 215 216 217 218 220 225 225 # Chol 226 230 230 230 230 231 232 232 232 234 234 236 236 238 240 243 # Chol 246 247 248 254 254 254 256 256 258 263 264 267 267 267 268 270 # Chol 270 272 278 278 283 285 300 300 308 327 334 336 353 393 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 10 11 12 13 14 15 16
The Heart Study began in 1948 by recruiting an Original Cohort of 5,209 men and women between the ages of 30 and 62 from the town of Framingham, Massachusetts, who had not yet developed overt symptoms of cardiovascular disease or suffered a heart attack or stroke. Assuming the above data came from healthy individuals then p95 = 334. This is way above current standards: Total Cholesterol Level Category Less than 200 MG/DL Desirable 200-239 MG/DL Borderline High 240 MG/DL and above High
Tests for Normality Tests for Normality Tests to Consider [1] Chi-Square Goodness-of-Fit Test [2] Graphics Normal Probability Plot [3] Non-Parametric Kolmogorov-Smirnov Test [4] Geary s Test for Normal Distribution [5] D Agostino-Belanger-Pearson s Skewness, Kurtosis, and Omnibus Tests [6] Others Shapiro-Wilk, Anderson-Darling, Cram r-von Mises Tests
Other considerations Other considerations Use of powers and other transforms Other distributions Fitting by first 4 moments (Pearson system) vs. Least Squares Confidence Intervals for predictions Jackknife method or Resampling/Bootstrapping to get confidence intervals
Mathematicians who contributed Mathematicians who contributed Karl Friedrich Gauss Joseph Gram Carl Charlier Karl Pearson William S. Gosset (Student) Norman Lloyd Johnson George Box David Cox John W. Tukey 1777-1855 1850-1916 1862-1934 1857-1936 1876-1937 1917-2004 1919-2000 1924- 1915-2000 Germany Denmark Sweden UK UK UK/USA UK/USA UK/USA USA
Dedicated to Prof. Horace F. Martin Dedicated to Prof. Horace F. Martin 1931-2010 B.S. M.S. Ph.D. M.D. J.D. M.P.H. - Providence College 1953 - University of Rhode Island 1955 - Boston University 1961 - Brown University 1975 - S. New England Sch. of Law 1990 - McGill University 2000 44
Thank You Florida Gulf Coast University 45