Methods for RNA Purification and Separation from DNA
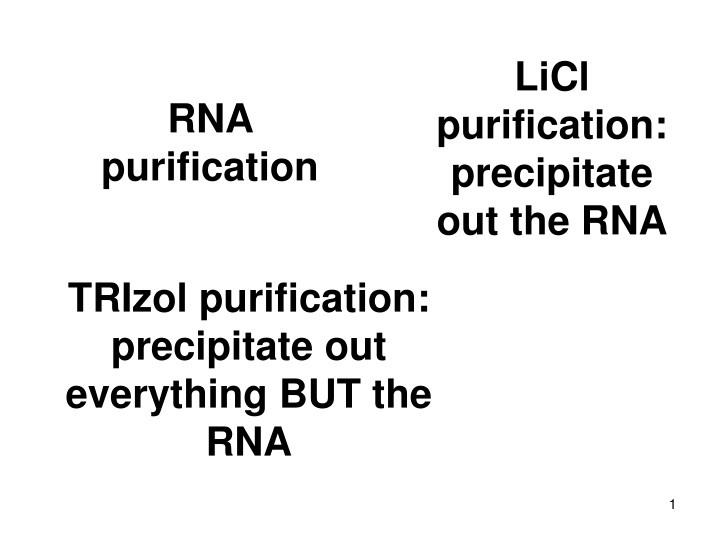
Selective precipitation of RNA using Lithium Chloride (LiCl) and extraction with phenol at acidic pH are two common methods for separating RNA from DNA. LiCl forms a complex with RNA facilitating its precipitation, while acidic phenol solution helps stabilize RNA at pH 4-5 for extraction. TRIzol variant includes guanidinium thiocyanate and chloroform for nucleic acid extraction, followed by salt precipitation. DNases are not commonly used for DNA removal from RNA due to low efficiency and cost.
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
You are allowed to download the files provided on this website for personal or commercial use, subject to the condition that they are used lawfully. All files are the property of their respective owners.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.
E N D
Presentation Transcript
LiCl RNA purification: precipitate out the RNA purification TRIzol purification: precipitate out everything BUT the RNA 1
Two common methods for separation of RNA from DNA 1. Selective precipitation of RNA using Lithium Chloride (LiCl) 2. Extraction with phenol buffered at an acidic pH (~4.5) NOTE: DNases are typically NOT used to remove large amounts of DNA from RNA because DNase is too expensive and is not efficient enough for that purpose Some protocols that have a small amount of DNA contamination might specify using DNase 2
Principle of Selective precipitation of RNA using Lithium Chloride (LiCl) Li+ ions form a Li+ RNA- complex with no net charge, which precipitates out of the solution Single stranded nature of RNA and the 2 OH group facilitate this The procedure is done using high concentration of LiCl and incubation on ice to promote the precipitation of the RNA salt Best for RNAs above 100 bases long 3
Selective precipitation of RNA using Lithium Chloride (LiCl) Add 7.5M LiCl to your crude lysate (having spun out the cell debris) Incubate at -20oC (i.e. in the freezer) for about 30 min (or longer) Centrifuge to collect the RNA pellet Desalt with very cold (-20oC) 70% ethanol (look at the LiCl concentration and consider the importance of this step) Resuspend in nuclease free water or other solution 4
Extraction with phenol solution buffered at acidic pH RNA is most stable at pH 4-5, DNA is most stable pH 8 Combine extraction buffer and organic solvent: Suspend cells, homogenized or powdered tissue in acidic phenol solution (phenol + water with buffer at acidic pH) Add chloroform, mix well, and centrifuge Separates into lower organic phase and upper aqueous Collect the upper - aqueous - layer that contains RNA May be a noticeable interphase between the two layers - avoid touching this with the pipette! 5
TRIzol variant of acidic buffered phenol, guanidinium thiocyanate, chloroform extraction Mix tissue powder with trizol and vortex Aqueous Centrifuge Collect aqueous phase add chloroform and shake DNA, protein, cell wall Organic isopropanol salt precipitation centrifugation Crude lysate containing nucleic acids and other cell constituents - proteins denature RNA pellet The aqueous phase contains water- soluble molecules, including RNA wash pellet with 70-85% ethanol dry pellet resuspend in nuclease free water Trizol (trireagent) monophasic solution of 1. guanidinium salt 2. phenol 3. buffer with low pH High-molecular weight DNA fragments and proteins in the interphase. RNA solution 6
Once you have an RNA solution, there is a risk of: Some RNase remaining (from the tissue originally) The solution: Common to add protein inhibitors of RNase RNase introduced (breath, reagents, equipment etc.) (RNAsin is one of them) Commercially available, but expensive Sensitive to denaturants (heat, chaotropic salts, phenol/chloroform Add only at end of procedure 7