Fungal Diversity Analysis in Wheat Roots Using Different Primers
Comparison of fungal taxonomic profiles in wheat roots using different primers, showcasing relative abundances of Glomeromycotina and Mucoromycotina orders in different treatments and varieties. Box plots demonstrate ASV evenness, observed ASV numbers, and Shannon diversity index, revealing insights into community diversity with respect to N treatments and wheat varieties.
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
You are allowed to download the files provided on this website for personal or commercial use, subject to the condition that they are used lawfully. All files are the property of their respective owners.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.
E N D
Presentation Transcript
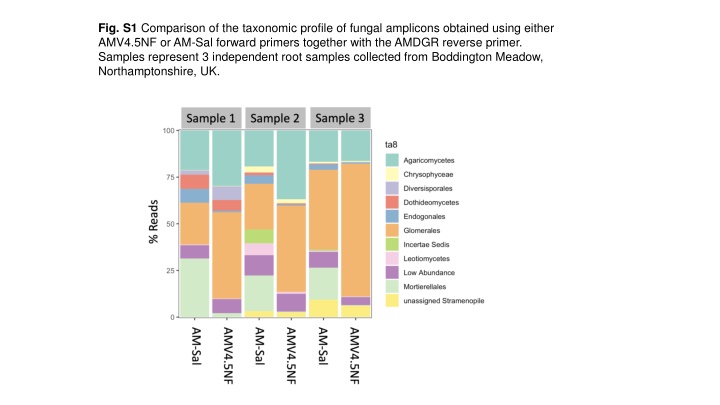
Fig. S1 Comparison of the taxonomic profile of fungal amplicons obtained using either AMV4.5NF or AM-Sal forward primers together with the AMDGR reverse primer. Samples represent 3 independent root samples collected from Boddington Meadow, Northamptonshire, UK.
Fig. S2 Relative abundances of Glomeromycotina and Mucoromycotina orders in roots of Aszita and Skyfall wheat varieties in zero-N and mineral-N treatments. Data was obtained using AM primers (AM-Sal-F forward and AMDGR reverse primers).
Fig. S3 Box plot showing ASV evenness, number of observed ASV and Shannon diversity index of combined Glomeromycotina and Mucoromycotina communities. Sequencing was performed using AM primers (AM-Sal-F forward and AMDGR reverse primers. A) Comparison of mineral-N and zero-N treatments B) Comparison of wheat variety. Numbers indicate p-values for pairwise comparison by Kruskal- Wallis test (ns = not significant, * 0.05, ** 0.01, *** 0.001).
Fig S4 Box plot showing ASV evenness, number of observed ASV and Shannon diversity index of Glomeromycotina communities. Sequencing was performed using AM primers (AM-Sal-F forward and AMDGR reverse primers. A) Comparison of mineral-N and zero-N treatments B) Comparison of wheat variety. Numbers indicate p-values for pairwise comparison by Kruskal-Wallis test (ns = not significant, * 0.05, ** 0.01, *** 0.001).
Fig S5 Box plot showing ASV evenness, number of observed ASV and Shannon diversity index of Mucoromycotina communities. Sequencing was performed using AM primers (AM-Sal-F forward and AMDGR reverse primers. A) Comparison of mineral-N and zero-N treatments B) Comparison of wheat variety. Numbers indicate p-values for pairwise comparison by Kruskal-Wallis test (ns = not significant, * 0.05, ** 0.01, *** 0.001).