Enzymes: Nature's Biochemical Catalysts
Enzymes are highly specific biological catalysts essential for body reactions. Catalyzing the conversion of compounds, they lower activation energy, speeding up reactions without being consumed. Each enzyme has recommended and systematic names, following clear nomenclature by the International Union of Biochemists. Enzymes are classified into classes based on their functions, with names reflecting the type of reaction they catalyze.
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
You are allowed to download the files provided on this website for personal or commercial use, subject to the condition that they are used lawfully. All files are the property of their respective owners.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.
E N D
Presentation Transcript
ENZYMES : are effective & high specific biological catalyst that catalyze the conversion of one or more compounds (substrates) into one or more different compounds (products). enzymes are involved in all essential body reactions & found in all body tissues Enzymes = Life
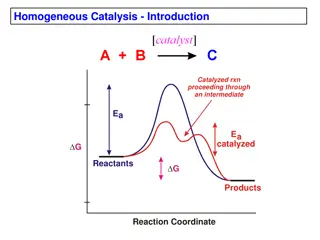
Catalyst is a substance that causes or accelerate a chemical reaction without itself being affected . With a catalyst, reactions occur faster and with less energy. Like all catalysts, enzymes are neither consumed nor permanently altered as a consequence of their participation in a reaction.
Enzymes are proteins that increase the reaction by lowering the energy of activation rate of Like enzymes lower activation energy by bringing the reactants closer together, aligning reactants, and/or weakening bonds so proceed faster. other catalysts, chemical reactions
Each enzyme is assigned two names: 1. The first is its short, Recommended Name, convenient for everyday use. 1. The second is the more complicated Systematic Name, which is used when an enzyme must be identified without ambiguity.
Recommended Name : The commonly used names for most enzymes describe the type of reaction catalyzed, followed by the suffix -ase. For example: dehydrogenases remove hydrogen atoms proteases hydrolyze proteins isomerases catalyze rearrangements in configuration. Modifiers may precede the name to indicate: the substrate (xanthine oxidase) the source of the enzyme (pancreatic ribonuclease) its regulation (hormone-sensitive lipase) Alphanumeric designators are added to identify multiple forms of an enzyme (eg, RNA polymerase III; protein kinase C ).
The International Union of Biochemists (IUB) developed a clear system of enzyme nomenclature in which each enzyme has a unique name and code number that identify the type of reaction catalyzed and the substrates involved. Despite the clarity of the IUB system, the names are relatively difficult, so we generally continue to refer to enzymes by their traditional names. In this system enzymes are grouped into the following six classes.
1. Oxidoreductases: Enzymes that catalyze oxidations and reductions. 2. Transferase: Enzymes that catalyze transfer of moieties such as glycosyl, methyl, or phosphoryl groups. 3. Hydrolases: Enzymes that catalyze hydrolytic cleavage of C C, C O, C N and other covalent bonds. 4. Lyases: Enzymes that catalyze cleavage of C C, C O, C N and other covalent bonds by atom elimination, generating double bonds. 5. Isomerases: Enzymes that catalyze geometric or structural changes within a molecule. 6. Ligases: Enzymes that catalyze the joining together (ligation) of two molecules in reactions coupled to the hydrolysis of ATP.
The IUB name for hexokinase illustrates both the clarity of the IUB system and its complexities. The IUB name of hexokinase is : ATP:D-hexose 6-phosphotransferase E.C.2.7.1.1. This name identifies hexokinase as a member of class 2 (transferases) Subclass 7 (transfer of a phosphoryl group) Subclass 1 (alcohol is the phosphoryl acceptor) Subclass 1 ( indicates that the alcohol phosphorylated is on carbon six of a hexose)
In addition to the previous nomenclature systems Some enzymes retain their original trivial names, which give no hint of the associated enzymic reaction, for example, trypsin and pepsin
Definitions and Related Terms 1- Holoenzyme & Apoenzyme: Some molecules other than protein for enzymic activity enzymes need Holoenzyme complete, enzyme with its non protein components . refer to the catalytically active Apoenzyme (zymogen) is the protein portion of a holoenzyme (i.e. the enzyme yet in an inactive form)
Many enzymes contain small non protein molecules & metal ions that participate directly in substrate binding or in catalysis. These termed prosthetic groups, cofactors, and coenzymes, these extend the catalytic capabilities of the enzymes.
Prosthetic groups are tightly and stably incorporated into a protein's structure by covalent or non covalent forces. Examples include : Pyridoxal phosphate, Flavin mononucleotide (FMN) & flavin adenine dinucleotide (FAD), Thiamin pyrophosphate, & biotin. Metal ions of (Co, Cu, Mg, Mn, and Zn.) Metal ions are the most common prosthetic groups. About one-third of all enzymes that contain tightly bound metal ions are termed metalloenzymes. Metals also may facilitate the binding and orientation of substrates & the formation of covalent bonds with reaction intermediates, or by acting as Lewis acids or bases to render substrates more electrophilic (electron-poor) or nucleophilic (electron-rich), and hence more reactive.
Cofactors serve functions similar to those of prosthetic groups, but bind in a transient, dissociable manner either to the enzyme or to a substrate, e.g. ATP. Unlike the stably associated prosthetic groups, cofactors must be present in the medium surrounding the enzyme for catalysis to occur. The most common cofactors also are metal ions. Enzymes that require a metal ion cofactor are termed metal-activated enzymes to distinguish them from the metalloenzymes for which metal ions serve as prosthetic groups.
Coenzymes serve as recyclable shuttlesor group transfer agents that transport many substrates from one point within the cell to another. The function of these shuttles is : First: They stabilize species such as hydrogen atoms (FADH) or hydride ions (NADH) that are too reactive to persist for any significant time period in the presence of the water or organic molecules within cell interior. Second:They also serve as an adaptor or handle that facilitates the binding of small chemical groups, such as acetate (coenzyme A) by their target enzymes. Other chemical moieties transported by coenzymes include methyl groups (folates) .
The active site is a region within an enzyme that fits the shape of substrate molecules. Amino acid side-chains align to bind the substrate through H- bonding, salt-bridges, hydrophobic interactions, etc. Products are released when the reaction is complete (they no longer fit well in the active site) and
Allosteric Site: Non-active site May interact with other substances resulting in overall enzyme shape change & inhibition or regulation.
When a substrate (S) fits properly in an active site, an enzyme-substrate (ES) complex is formed: E + S ES Within the active site of the ES complex, the reaction occurs to convert substrate to product (P): ES E + P The products are then released, allowing another substrate molecule to bind the enzyme - this cycle can be repeated millions (or even more) times per minute The overall reaction for the conversion of substrate to product can be written as follows: E + S ES E + P
The reaction for the sucrase catalyzed hydrolysis of sucrose to glucose and fructose can be written as follows: E + S ES E + P1 + P2 where E = sucrase, S = sucrose, P1 = glucose and P2 = fructose
Two models are proposed : 1- The Lock-and-Key Mode 2- The Induced Fit Model
In the lock-and-key model of enzyme action: - The active site has a rigid shape - Only substrates with the matching shape can fit - The substrate is a key that fits the lock of the active site This is an older model, however, and does not work for all enzymes
In the induced-fit model of enzyme action: - The active site is flexible, not rigid - The shapes of the enzyme, active site, and substrate adjust to maximumize the fit, which improves catalysis - There is a greater range of substrate specificity This model is more consistent with a wider range of enzymes
While Fischer's "lock and key model" accounted for the grate specificity of enzyme-substrate interactions, the implied rigidity of the enzyme's active site failed to account for the dynamic changes that we now know accompany catalysis. This was addressed by Daniel Koshland's induced fit model, which states that: When substrates approach and bind to an enzyme they induce a conformational change analogous to placing a hand (substrate) into a glove (enzyme) The enzyme in turn induces reciprocal changes in its substrates, harnessing the energy of binding to facilitate the transformation of substrates into products.
Enzymes use various combinations of four mechanisms to catalytize a chemical reactions these are: Catalysis by Proximity Acid Base Catalysis Catalysis by Strain Covalent Catalysis 1. 2. 3. 4.
Isoenzymes are different forms of an enzyme that catalyze the same reaction in different tissues in the body. They have slight variations in the amino acid sequences of the subunits of their quaternary structure Different isoenzymes may arise from different tissues and their specific detection may give clues to the site of pathology.
Pyruvate Lactate (anaerobic glycolysis) lactate dehydrogenase (LDH), which converts lactate to pyruvate, during anaerobic glycolysis It is a tetrameric protein and made of two types of subunits namely H = Heart, M = skeletal muscle It exists as 5 different isoenzymes with various combinations of H and M subunits LDH is elevated in myocardial infarction, blood& liver disorders
LACTATE DEHYDROGENASE ISOENZYMES
Creatine + ATP phosphocreatine + ADP (Phosphocreatine serves as energy reserve during muscle contraction) Creatine kinase is a dimer made of 2 monomers M subunit, & B subunits Three different isoenzymes are formed
Isoenzyme Name Composition Present in Elevated in CK-1 BB Brain CNS diseases Acute myocardial infarction Myocardium / Heart CK-2 MB Skeletal muscle, Myocardium CK-3 MM
The ability to assay the activity of specific enzymes in blood aids in the diagnosis and prognosis of disease. Deficiencies in the quantity or catalytic activity of key enzymes can result from genetic defects, nutritional deficits, or toxins. The absolute specificity of enzymes is of a particular value for using them as catalysts for specific reactions in the synthesis of a drug or antibiotic.
Enzymes also can be employed in the clinical laboratory as tools for determining the concentration of critical metabolites. For example, glucose oxidase is frequently utilized to measure plasma glucose concentration. Enzymes are also used for the treatment of injury and disease. Tissue plasminogen activator (tPA) or streptokinase is used in the treatment of acute MI, while trypsin has been used in the treatment of cystic fibrosis
Presence of disease Organs involved Aetiology /nature of disease: differential diagnosis Extent of disease-more damaged cells-more leak enzymes in blood Time course of disease