DNA Modifying Enzymes
DNA modifying enzymes play crucial roles in various DNA metabolic processes such as replication, recombination, and repair. Nucleases are key enzymes capable of cleaving phosphodiester bonds in nucleic acids, essential for DNA and RNA processing and degradation. Exonucleases remove nucleotides from DNA strands, while endonucleases act within target molecules. Defective nuclease activities can lead to autoimmune diseases. Explore the functions and importance of nucleases in DNA and RNA metabolism.
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author. Download presentation by click this link. If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
E N D
Presentation Transcript
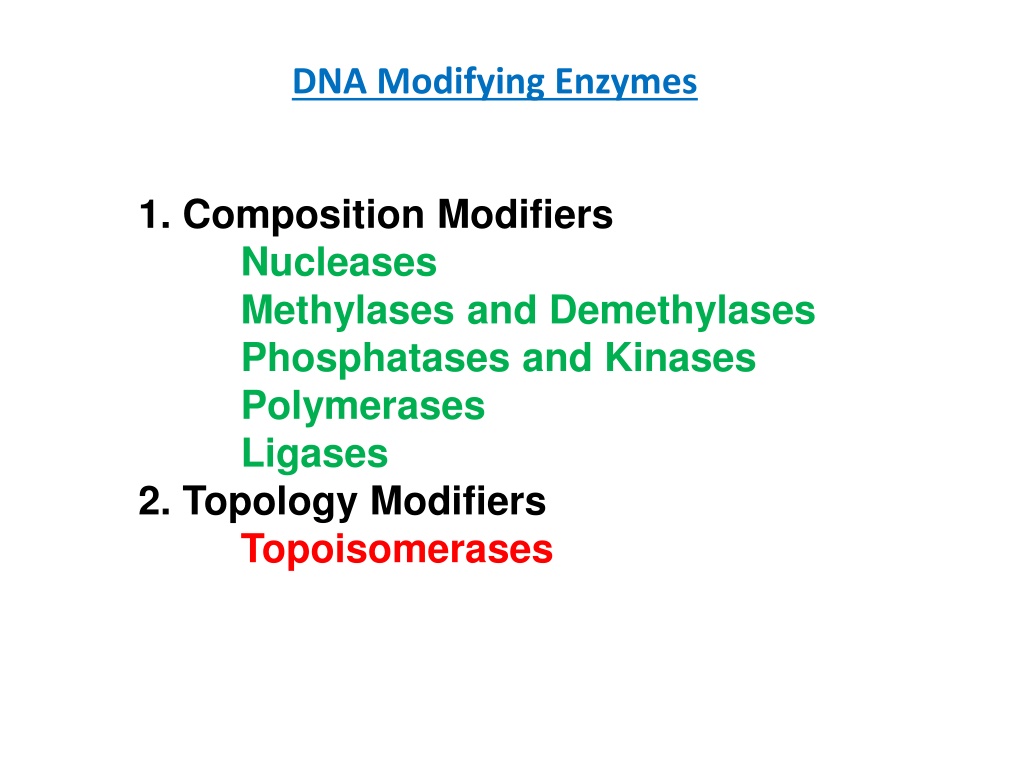
DNA Modifying Enzymes 1. Composition Modifiers Nucleases Methylases and Demethylases Phosphatases and Kinases Polymerases Ligases 2. Topology Modifiers Topoisomerases
Nucleases: A nuclease is an enzyme capable of cleaving the phosphodiester bonds between nucleotides of nucleic acids. Exonucleases digest nucleic acids from the ends. Endonucleases act on regions in the middle of target molecules. They are further subcategorized as deoxyribonucleases and ribonucleases. The former acts on DNA, the latter on RNA. Nuclease activities are integral parts of DNA replication; the 5 to 3 exo- and endonucleases are needed to remove RNA primers and the 3 to 5 exonuclease for proofreading. Two other major DNA metabolic processes, recombination and repair, are initiated by nucleases. Nuclease activity is also required for structural alterations of nucleic acids, for example, topoisomerization site-specific recombination and RNA splicing during which a phosphodiester bond is temporarily broken and reformed after strand passing or transfer to a new target.
In addition, nuclease activities are essential in RNA processing, maturation, and RNA interference. RNA and DNA degradation is an essential component of microbial defense mechanisms. Nucleases are even essential for programmed cell death . Defective DNase and RNase activities have been associated with various autoimmune diseases due to incomplete removal of endogenously produced nucleic acids A nuclease is a phosphodiesterase that cleaves one of the two bridging P-O bonds, 3 or 5 , in a nucleic acid polymer
Exonucleases, which remove one nucleotide at a time from the end of a strand, can be further divided to two groups by the 5 to 3 versus 3 to 5 polarity.
Exonuclease usually attack either the 5 or 3 ends. 1. Source: Escherichia coli Exonuclease I breaks apart single-stranded DNA in a 3' 5' direction, releasing deoxyribonucleoside 5'-monophosphates one after another. ExoI ssDNA 2. Source: Escherichia coli Exonuclease III breaks apart double-stranded DNA in a 3' 5' direction, releasing deoxyribonucleoside 5'-monophosphates one after another. ExoIII dsDNA
3. WRN Exonuclease Werner Syndrome>>>>>>autosomal recessive disorder 3' 5' direction dsDNA 4. TREX1 and TREX2 are the major mammalian 3 5 exonucleases and also prefer single stranded DNA substrate .. ssDNA Aicardi-Gouti res syndrome, a disorder that involves severe brain dysfunction (encephalopathy) >>>>>>>>>>>autosomal recessive disorder 5. RNaseT 3' 5' direction ssRNA 6. 5 -3 exoribonuclease 1 (Xrn1) is a protein that in humans is encoded by the XRN1 gene. Xrn1 hydrolyses RNA in the 5 to 3 direction.
>>>>>>>>>>>>>Nuclease BAL31 exonuclease degrades both 3' and 5' termini of duplex DNA without generating internal scissions. BAL 31 Nuclease acts as an exonuclease, degrading double-stranded DNA and RNA from both 5'-phosphate and 3'-hydroxyl termini. This enzyme also possesses single- stranded DNA and RNA endonuclease activity and is capable of cleaving at DNA/RNA nicks and gaps. It requires Ca2+as an essential cofactor for its exonuclease and endonuclease activities. Source: Alteromonas espejiana
Members of the Flap endonuclease 1 (FEN1) family have the 5 to 3 exonuclease activity in addition to the endonuclease activity. Mre11, which is involved in DNA double-strand break (DSB) repair, has both endo and 3 to 5 exo-nuclease activities Q1. Can exonuclease cut plasmid?
Endonucleases >>>Break internal phosphodiester bonds A. Mung Bean Nuclease: is a nuclease derived from sprouts of the mung bean (Vigna radiata). >Single-stranded (ssDNA or RNA) specific endonuclease > Catalyzes the removal of single-stranded extension in double-stranded DNA Mung Bean can be used for: 1. Removal of both 3' and 5' single-stranded overhangs from double-stranded DNA to create blunt ends 2. Cleavage of single-stranded DNA and RNA 3. Cleavage of the single-stranded region in a DNA hairpin 4. Remove ssDNA from a mixture also containing dsDNA Mung bean nuclease requires Zn2+. The addition of EDTA or SDS causes irreversible inactivation.
B. Nuclease S1 Aspergillus oryzae S1 Nuclease (Zinc dependent ) degrades single-stranded nucleic acids (ssDNA or RNA), releasing 5'-phosphoryl mono- or oligonucleotides. It is five times more active on DNA than on RNA. S1 Nuclease can introduce breaks into double-stranded DNA, RNA and DNA/RNA hybrids at high enzyme and low salt concentrations. Applications: 1. Removal of single-stranded overhangs of DNA fragments 2. S1 transcript mapping 3. Cleavage of hairpin loops Blunt Ends Nuclease P1 found in Penicillium citrinum Nuclease protection assays
>Finding the Transcription Start Site (TSS) >Also Intron Site
RNase A degrades single-stranded RNA at C and U residues. RNase H (Ribonuclease H) is an endoribonuclease that specifically hydrolyzes the phosphodiester bonds of RNA which is hybridized to DNA. This enzyme does not digest single or double-stranded DNA. It can be used remove mRNA during second strand cDNA synthesis. DNaseI
Alkaline Phosphatase : Remove phosphate group fron the 5 end of the DNA molecule Source: E. coli or Calf Intestinal Tissue
T4 Polynucleotide kinase Addition of Phosphate Group at the 5 end 5' phosphorylation of DNA/RNA for subsequent ligation
Terminal Deoxynucleotidyl Transferase (TdT) Terminal transferase catalyzes the addition of deoxynucleotides to the 3' hydroxyl terminus of DNA molecules. Homopolymeric tailing of linear duplex DNA with any type of 3'-OH terminus Homopolymer tailing is the way for the ligation of blunt ended DNA fragments. tailing involves using an enzyme terminal deoxynucleotidyl transderase. This enzyme will repeadly add nucleotide to the 3` end of the double stranded DNA molecule