DNA, Chromosomes, and Chromatin Structure
DNA is made up of genes, chromosomes, and chromatin. Genes carry vital information for protein synthesis, while chromosomes are condensed DNA required for cell division. Junk DNA are non-coding regions, and sister chromatids are identical DNA copies. Homologous chromosomes have matching structures, and packaging involves DNA-protein associations forming chromatin. The chromatin structure allows DNA replication and transcription through nucleosomes and chromatosomes.
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
You are allowed to download the files provided on this website for personal or commercial use, subject to the condition that they are used lawfully. All files are the property of their respective owners.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.
E N D
Presentation Transcript
Gene It is a segment of DNA that contains information for synthesizing proteins or RNA. Hence, it is part of DNA that is carrying the vital information. Chromosome The chromosome is the most compact and condensed form of DNA. The chromosome form of DNA is required for its equal partition into daughter cells. This condensation is accomplished by folding the chromatin fibers into loops, coils and supercoils. To understand the extent of packaging and coiling, imagine a rope of length equal to the field length of the football playing ground and this rope is coiled and condensed to half an inch ball. Junk DNA The segments or parts of DNA which do not carry any information of synthesizing protein or RNA is called junk DNA. It is non coding region. Such junk DNA are interspersed between the genes. It acts like spaces and its true function is yet to be discovered clearly.
Sister Chromatids- It is the identical copies of the DNA formed after the DNA replication i.e. in . The replicated copies are held together by a common centromere. Haploid cell The cell carrying a single set of chromosomes. Ex. haploid human cells like egg and sperm have 23 chromosomes. Diploid cell The cell carrying a double set of chromosomes. Ex. diploid human somatic cells have 23 + 23 chromosomes = 46.
Homologous chromosomes In diploid cells, there are two sets of chromosomes. During sexual reproduction, the offspring receives one set of chromosome from both the parents (one is paternal and other is maternal). The chromosomes from both parental sets that have the same structure, size, gene loci and banding pattern are called homologous chromosomes. Heterologous chromosomes The chromosomes that do not have the same structure, size, gene loci and banding pattern as called heterologous chromosomes.
Packaging of Chromosomes The DNA present in the nucleus is always found to be associated with proteins. This DNA- protein association is called chromatin. This is a less condensed form of DNA. It is found in the interphase of the eukaryotic cell cycle, when the cell is not dividing. The chromatin structure allows the DNA replication and transcription. Under electron microscope, the chromatin structure looks like beads on string, where the DNA is tightly associated with histone proteins. The DNA is wrapped around the core of histone proteins, and it appears like a bead. These tiny beads are called nucleosomes and their diameter is around 11nm. While wrapping, 146 base pairs of the DNA show slightly less than two turns (1.7) around the histone core. Each histone core has two copies of H2A, H2B, H3 and H4 histone proteins hence forming histone octamer. The histone proteins are rich in positive amino acids and thus they allow negatively charged DNA to wrap around it. The DNA and histone proteins interact electrostatically and with non covalent bonds. When the nucleosomes are linked by an additional histone protein called H1 protein and linker DNA (20bp) it forms a chromatosome. Chromatosomes are the fundamental unit of chromatin structure. The core histone proteins N terminal tail extends out. The tails are subjected to covalent modification like acetylation, methylation or phosphorylation responsible for modelling and remodelling the chromatin structure.
The Formation of nucleosomes and chromosomes is the first stage of condensation. This reduces the length of DNA by sevenfold i.e. one meter DNA becomes 14cm long. But this is still not sufficient to get accommodated in a microscopic cell. Hence, the chromatin needs to fold to more extent. When chromatin is folded, packaged and condensed, it is called a chromosome. The structure of chromatin and chromosome is changeable and it is visible in different stages of the eukaryotic cell cycle.
The second level of compaction is 30 nm fiber because its diameter is 30nm. The beads on string folds in an irregular pattern forming 30nm fiber. The H1 protein plays a vital role in stabilizing 30 nm fiber. The 30nm fiber reduces the length of DNA by 50 times. The 30nm chromatin fiber behaves like a fluid mosaic showing different variations of the zig-zag model. In the zig zag model, the successive nucleosomes along with DNA are stacked alternatively. Such close proximity of nucleosomes favours interaction of adjacent nucleosomes s histone protein tail. The 30nm fiber is further folded to thick fibers forming 300nm to 700nm condensed chromatin loop and finally to 1400nm chromosome. This is achieved by forming chromatin loops around the scaffold protein. Such compaction, condenses the DNA from 1cm to 1um i.e. the packing ratio is 10,000:1.
Packaging of DNA molecules into chromosome
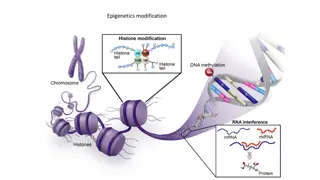
The compaction of DNA hampers the molecular process like replication and transcription. In order to process the molecular processes, the DNA needs to unwind from the histone octamer (nucleosome) and open the helix and expose the template to the enzymes. The unwinding of DNA is achieved by modification of amino acid groups like acetylation, methylation and phosphorylation of histone protein N terminal tails. The modification of tails destabilises the non-covalent bonds between DNA and histone proteins allowing the unwrapping of DNA. This is called modelling of chromatin. This process is reversible and allows remodelling of chromatin. The modelling and remodelling is an energy driven process. The packaging of DNA not only allows DNA to be accommodated in a small space but also permits gene expression. While looping, the related functional genes are brought together in close proximity to fulfil the need of the cell. While DNA packaging, the highly condensed chromatin region is called Heterochromatin and less condensed chromatin region is called Euchromatin. In Heterochromatin, the gene expression is inhibited and active gene expression is observed in euchromatin.
Acetylation: Transcriptional Activation Methylation is added to the lysine or arginine residues of histones H3 and H4, with different impacts on transcription. Arginine methylation promotes transcriptional activation while lysine methylation is implicated in both transcriptional activation and repression depending on the methylation site. Examples: Histone modification Function Location H3K4me1 Activation Enhancers H3K4me3 Activation Promoters, bivalent domains H3K9Ac Activation Enhancers, promoters H3K27me3 Repression Promoters in gene-rich regions, developmental regulators, bivalent domains H3K9me3 Repression Satellite repeats, telomeres, pericentromeres H4 Arg3 methylation Activation
The major categories of histone writers and erasers: Modification Writers Erasers Acetylation Histone acetyltransferases (HATs) Histone deacetylases (HDACs) Methylation Histone methyltransferases (HMTs/KMTs) and protein arginine methyltransferases (PRMTs) Lysine demethylases (KDMs) Phosphorylation Kinases Phosphatases