Detection Methods for GMOs and LMOs in Molecular Biology
Techniques in Molecular Biology lecture discusses GMOs and LMOs, transgenic plants, examples like Bt cotton and Golden rice, detection methods, purpose of detection, and how transgenic plants are created. The content emphasizes the need to differentiate GM crops from non-GM crops and the importance of accurate detection for various stakeholders.
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
You are allowed to download the files provided on this website for personal or commercial use, subject to the condition that they are used lawfully. All files are the property of their respective owners.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.
E N D
Presentation Transcript
Techniques in Molecular Biology 2017 Fall - Lecture -8 Detection methods for GMOs/LMOs by Jasmin Sutkovic , 14thDecember 2017
Definition GMO / LMO Any living organism that possesses a novel combination of genetic material obtained through the use of modern biotechnology Living organism means biological entity capable of transferring or replicating genetic material including sterile organisms, viruses and viriods
Transgenic plants Characterized by the insertion of a new gene or sets of genes into their genome The new genes translate and new protein expressed This gives the plant new characteristic
Transgenic plants - Examples Bt cotton Cotton plants resistant to lepidopteran insects Round-up ready soybean Soybean resistant to glyphosate Golden rice Rice grains with beta-carotene
Detection Methods GM crops are indistinguishable from non- GM crops to the naked eye ==
Detection Methods Basis of detection is to exploit the differences between the unmodified variety and the transgenic plant Testing methods need to look for the genes (DNA) introduced into the crop for the proteins produced in the plant by the introduced gene Workshop on Safety Assessment and Regulation of GM crops; 2-3 August 2006
Detection methods purpose Producers of LMO To assure purity and segregation of products To be able to trace genetic modification in breeding Food & feed industry, seed companies To assure purity and segregation of products To assure compliance with legislation Competent (enforcement) authorities Product control, compliance with legislation To be able to retrieve specific products e.g. if marketing permission withdrawn Workshop on Safety Assessment and Regulation of GM crops; 2-3 August 2006
How transgenic plants are made? Identification of useful gene The cloning of the gene into a suitable plasmid vector, Delivery of the vector into plant cell (insertion and integration) followed by expression and inheritance of the foreign DNA encoding a polypeptide. A gene construct consists typically of three elements: The promoter functions as an on/off switch for when and where the inserted/modified gene is active in the recipient plant The transgene encodes a specifically selected trait The terminator functions as a stop signal for transcribing the inserted/altered gene marker genes for distinguishing GM from non-GM cells during crop development Workshop on Safety Assessment and Regulation of GM crops; 2-3 August 2006
How transgenic plants are made? Promoter Gene Terminator transformation Plant DNA before transformation Plant DNA after transformation Workshop on Safety Assessment and Regulation of GM crops; 2-3 August 2006
General Procedure Detection: to determine whether a product is GM or not. For this purpose, a general screening method can be used. The result is a positive/negative statement. Identification: to find out which GM crop or product are present and whether they are authorized or not in the country. Quantification: If a crop or its product has been shown to contain GM varieties, then it become necessary to assess compliance with the threshold Regulation by the determination of the amount of each of the GM variety present.
Detection methods GM Detection DNABased methods Protein based methods To detect foreign DNA To detect novel protein Qualitative and Quantitative
Detection methods requirements Target molecules DNA or protein Capture molecules For DNA: primers and probes For proteins: antibodies Reference materials Positive and negative controls Calibrants for quantitation
Detection methods requirements Target molecules Degraded or absent target molecules undetectable! Effects of processing usually significant Important to improve methods to recover target Capture molecules Impossible without description of target molecules Need material for method development / validation
DNA based methods Highly sensitive - can detect trace amounts GM- DNA Work with most product types - both processed and unprocessed products Can test for multiple GM varieties simultaneously Takes a number of days to perform - typically 3-5 days Requires highly skilled personnel and laboratory analysis More expensive than Protein Based Methods
Protein based methods Relatively cheap to perform Rapid turnaround - 5 to 20 mins for strips; 24 hours for ELISA Strips do not require trained personnel Limited to one or a small number of varieties per test Not appropriate for processed products Not appropriate for some GM varieties - in certain crops the GM protein is only produced in the leaves or stems and not in the actual grain. Protein tests on the grain are therefore not informative. Not very sensitive (~1% of GM protein)
Protein based methods ELISA Lateral flow strip
Protein based methods Enzyme-Linked Immunosorbent Assay (ELISA) Enzyme immunoassay utilizing an enzyme-labelled immunoreactant (antigen or antibody) and immunosorbent (antigene or antibody bound to solid support) Enzyme catalyses colour reaction more intense colour, more proteins present Quantification based on standard curve (reference)
Protein based methods Enzyme-Linked Immunosorbent Assay (ELISA) Prepare a surface to which a known quantity of antibody is bound. Apply the antigen-containing sample to the plate. Wash the plate, so that unbound antigen is removed. Apply the enzyme-linked antibodies which are also specific to the antigen. Wash the plate, so that unbound enzyme-linked antibodies are removed. Apply a chemical which is converted by the enzyme into a fluorescent signal. View the result: if it fluoresces, then the sample contained antigen 1. 2. 3. 4. 5.
Protein based methods Enzyme-Linked Immunosorbent Assay (ELISA)
Protein based methods Enzyme-Linked Immunosorbent Assay (ELISA) Specific Antibodies Positive control Negative control Buffer control Each sample in duplicate wells
Protein based methods Lateral flow stick
Protein based methods Lateral flow stick Collect leaf and extract sample Place the strip into the extraction tube The sample will travel up the strip Allow the strip to develop for 10 minutes To retain the strip, cut off and Discard bottom section of the strip
Protein based methods Lateral flow stick
Protein based methods Lateral flow stick control test negative positive
Protein based methods Company Test Kit Test Format ELISA EnviroLogix Inc., USA Cry 1Ab & Cry1Ac in Bt corn and cotton Cry2A, Cry1C, Cry1F, Cry3B and Cry9C Cry9C ELISA Quickstix (1kernel in 800 in 5 min.) Lateral flow assay (Soybeans 0.1%; corn 0.125%) Lateral flow assay (corn 0.125%) ELISA Neogen Corporation, USA CP4 Cry9C Strategic Diagnostics Inc., USA RUR Soya Meal, Soya Grain, Bt 1 Maize, Bt 9 Maize
Protein based methods Advantages of ELISA Reasonably sensitive Less susceptible to false positives Low per sample cost Handles large number of samples Can be subjected to automation Detection kits available commercially
Protein based methods Disadvantages of ELISA High development costs for the assays GM products might be produced only during certain developmental stages or in certain plant parts and such GMs are difficult to be detected by ELISA In processed foods the proteins denatures easily, which makes it difficult to use ELISAfor processed food fractions
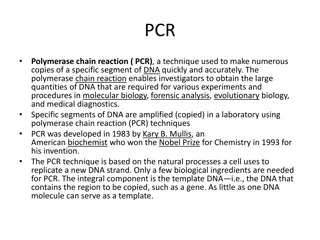
DNA based methods DNA based methods are based on detection of the specific genes, or DNA genetically engineered into the crop. Commercial testing is conducted using PCR technology The PCR technique is based on multiplying a specific target DNA allowing the million or billion fold amplification by two synthetic oligonucleotide primers.
DNA based methods PCR based detection Detects a known DNA sequence by amplification Target sequence is duplicated billions of times Results obtained can be quantitative or qualitative Enzyme :Taq polymerase Equipment : Thermocycler/PCR machine
DNA based methods Principles of PCR Denaturation primer annealing amplification 30 cycles
DNA based methods Sample homogenization DNAextraction Check for availability of Plant DNA Plant DNApresent Plant DNAabsent Screening PCR GMO present GMO absent GMO detection GMO identification
DNA based methods InsertDNA PlantDNA PlantDNA 1. Screening GMO Detection 2. Construct specific-PCR GMO identification 3. Event-specific PCR
DNA based methods Construct specific PCR gene x gene y gene z M P 1 2 3 1 2 3 1 2 3 1. Event 1 2. Event 2 3. Event 3
DNA based methods PCR Quality Assurance Positive and negative controls Extraction controls (Blanks) Validated primers All analysis performed at least in duplicate Internal/ external validation trials
DNA based methods Advantages of PCR High sensitivity. Detection limits 0.1% of DNA present Semi-quantitative PCR can provide a rough estimate of the level of contamination Real-Time PCR providing quite accurate quantitation in the less than 10% range DNA highly stable
DNA based methods Limitations of PCR Not all tissues contain the same amount of DNA Require relatively advanced lab. facilities and instrumentation and highly trained staff Cost is fairly high Time (usually >24 hrs for results) Special precautions need to be taken to avoid the contamination
DNA based methods Types of analysis Event specific quantification Quantification Identification Detection 1 2 3 4 Types of samples 1 2 3 4 grains, seeds Crudely processed material pure Crudely processed material mixed Processed products
DNA based methods M 1 2 3 4 5 Multiplex PCR-based detection methods 500bp 400bp 300bp 356bp fragment of EPS synthase gene 200bp 195bp fragment of 35S promoter 100bp Multiplex PCR for detection of 35S promoter and EPS synthase gene sequence in transgenic soybean
DNA based methods Quantitative PCR End point analysis (Competitive PCR) Real time analysis Micro-array Automated rapid screening Based on nucleic acid hybridisation Specific probes are attached to the chip LMO chip GeneScan Europe