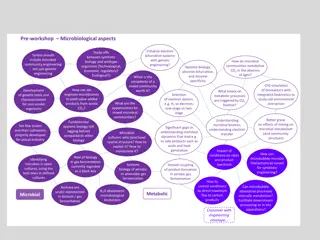
MicrobeDB Database Analysis for Microbial Diversity
In June 2017, data from MicrobeDB.jp was analyzed, revealing microbial diversity in various phyla and families. The study included Wilcoxon P-values and abundances of different microbial genera. The analysis indicated the presence of various bacterial and archaeal species in different environments, including biofilms, planktonic cells, and waste treatment systems. The findings provide insights into the composition and distribution of microbes in natural and artificial environments.
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
You are allowed to download the files provided on this website for personal or commercial use, subject to the condition that they are used lawfully. All files are the property of their respective owners.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.
E N D
Presentation Transcript
12. Jun. 2017 MicrobeDB.jp
Genus Family Phylum Wilcoxon_P-value Bio Pla abundance Rhodocyclaceae Methanomicrobiaceae Burkholderiales Comamonadaceae Comamonadaceae Nitrosomonadaceae Comamonadaceae Chitinophagaceae Cytophagaceae Alteromonadaceae Comamonadaceae Comamonadaceae Comamonadaceae Synergistaceae Haloarculaceae Rhodobacteraceae Nostocaceae Alcanivoracaceae Flavobacteriaceae - Comamonadaceae Borreliaceae Methylophilaceae Aeromonadaceae Legionellaceae Methanoregulaceae Halobacteriovoraceae Leuconostocaceae Thermodesulfobacteriaceae Veillonellaceae Firmicutes Dechloromonas Methanoculleus * Leptothrix Verminephrobacter Comamonas Nitrosomonas Rhodoferax Niastella Runella Marinobacter Acidovorax Polaromonas Delftia Acetomicrobium Haloarcula * Ketogulonicigenium Nostoc Alcanivorax Flavobacterium Gamma proteobacterium Variovorax Borrelia Methylotenera Oceanimonas Legionella Methanosphaerula * Halobacteriovorax Oenococcus Thermodesulfobacterium Veillonella *:Archaea Proteobacteria Euryarchaeota Proteobacteria Proteobacteria Proteobacteria Proteobacteria Proteobacteria Bacteroidetes Bacteroidetes Proteobacteria Proteobacteria Proteobacteria Proteobacteria Synergistetes Euryarchaeota Proteobacteria Cyanobacteria Proteobacteria Bacteroidetes Proteobacteria Proteobacteria Spirochaetes Proteobacteria Proteobacteria Proteobacteria Euryarchaeota Proteobacteria Firmicutes Thermodesulfobacteria 0.0727272727272727 0.104984533136531 0.109090909090909 0.109090909090909 0.109090909090909 0.163636363636364 0.163636363636364 0.163636363636364 0.163636363636364 0.163636363636364 0.163636363636364 0.163636363636364 0.163636363636364 0.215084048647664 0.218253353455316 0.23030303030303 0.23030303030303 0.23030303030303 0.23030303030303 0.23030303030303 0.23030303030303 0.23030303030303 0.23030303030303 0.23030303030303 0.23030303030303 0.287391141675814 0.294186288800238 0.294186288800238 0.297517091364237 0.315151515151515 -1.107110373 0.731165913 -0.835147277 -0.782962845 -0.759719236 -0.72715939 -0.799247961 -1.272785119 -1.134469589 -0.877247817 -0.737605993 -0.832885774 -0.773168569 -0.590426255 -1.178457194 -0.618396351 -1.232093037 -1.047552249 -0.93423204 -1.206114006 -1.162830868 -0.977969917 -0.551805721 -0.027911397 -0.309593169 -0.643086887 -0.802827733 -0.728746771 -0.774661106 -0.668095291 Bio Pla Pla majority Pla Bio
Wilcoxon / , / Bio Pla 1 Wilcoxon Pla 1 Pla Fisher Pla ? / Bio Bio count Pla Pla abundance Biofilm Pla bio B:104/224,P:258/272 Pla Bio DB Pla count Cutoff t Pla Bio / ? Fisher matrix odds
MicrobeDB.jp Biofilm Planktonic ? Waste treatment,food,artificial natural environment,fungi associated,large-scale artifact biofilm cells environment Biofilm,sediment,rock sand and soil,animal associated,plant associated,geographic feature and biome planktonic cells environment Water,aerobic environment,aquatic feature and biome,ice, Biofilm cell Planktonic cell
Planktonic Biofilm Biofilm Planktonic Wilcoxon : Fisher : KO : KO Biofilm
Genus Family Phylum Wilcoxon_P-value Nostocaceae - Legionellaceae Cytophagaceae Yersiniaceae Flavobacteriaceae Aeromonadaceae Ferrimonadaceae Rhodocyclaceae Crocinitomicaceae Sphingomonadaceae Nitrosomonadaceae Rhodobacteraceae Chitinophagaceae Cytophagaceae Eubacteriaceae Acidothermaceae Haliscomenobacteraceae Halomonadaceae Chitinophagaceae Pasteurellaceae Cytophagaceae Piscirickettsiaceae Alcaligenaceae Aeromonadaceae Methanomicrobiaceae Leuconostocaceae Merismopediaceae Jonesiaceae Sphingobacteriaceae Nostoc Gamma proteobacterium Legionella Leadbetterella Yersinia Flavobacterium Aeromonas Ferrimonas Dechloromonas Fluviicola Zymomonas Nitrosomonas Ketogulonicigenium Neisseria Runella Eubacterium Acidothermus Haliscomenobacter Halomonas Niastella Actinobacillus Flexibacter Methylophaga Pusillimonas Oceanimonas Methanoculleus (Archaea) Oenococcus Synechocystis Jonesia Pseudopedobacter Cyanobacteria Proteobacteria Proteobacteria Bacteroidetes Proteobacteria Bacteroidetes Proteobacteria Proteobacteria Proteobacteria Bacteroidetes Proteobacteria Proteobacteria Proteobacteria Bacteroidetes Bacteroidetes Firmicutes Actinobacteria Bacteroidetes Proteobacteria Bacteroidetes Proteobacteria Bacteroidetes Proteobacteria Proteobacteria Proteobacteria Euryarchaeota Firmicutes Cyanobacteria Actinobacteria Bacteroidetes 0.00606060606060606 0.00606060606060606 0.00606060606060606 0.00606060606060606 0.00606060606060606 0.00606060606060606 0.0121212121212121 0.0242424242424242 0.0242424242424242 0.0242424242424242 0.0242424242424242 0.0242424242424242 0.0242424242424242 0.0242424242424242 0.0242424242424242 0.0424242424242424 0.0424242424242424 0.0424242424242424 0.0424242424242424 0.0424242424242424 0.0424242424242424 0.0424242424242424 0.0424242424242424 0.0424242424242424 0.0424242424242424 0.0452199741787572 0.0700049832291192 0.0727272727272727 0.0727272727272727 0.0727272727272727
(Bio:224Pla:272) Genus Bio Pla Acaryochloris 88 240 136 32 Acetobacter 93 242 131 30 Acetobacterium 86 215 138 57 Acetohalobium 86 216 138 56 Acholeplasma 87 238 137 34 Achromobacter 102 257 122 15 Acidaminococcus 88 237 136 35 Acidianus 40 205 184 67 Acidilobus 64 203 160 69 Acidimicrobium 96 240 128 32 Acidiphilium 96 253 128 19 Pseudomonas Leptospira Escherichia 103 258 121 14 Fervidicoccus 28 210 196 62 Coprothermobacter Marinomonas 97 258 127 14 102 258 122 14 14 196 210 76 58 238 166 34
(Bio:224Pla:272) 2 Phylum Genus P-value (Fisher s test) Odds ratio(Bio > 1,Pla < 1) Spirochaetes Leptospira Fervidicoccus (Archaea) Coprothermobacter Halogeometricum (Archaea) Flavobacteriaceae Trichodesmium Leuconostoc Ferroglobus (Archaea) Thermobaculum Pelagibacterium Methanocella (Archaea) Staphylothermus (Archaea) Methylomonas Roseobacter Bdellovibrio Magnetococcus Halorubrum (Archaea) Parvularcula Parachlamydia Methanoregula (Archaea) Marinomonas Pseudoalteromonas Acidianus (Archaea) Prochlorococcus 1.84984756776945e-55 1.30666651736405e-50 1.2144453370268e-46 1.04875444172146e-45 7.15629803877711e-45 1.11039259592354e-44 2.57279815069165e-44 1.40508206857176e-43 2.25857826588668e-43 1.26051550425212e-42 9.15759027518411e-42 1.08395647986535e-41 1.35143151367562e-41 1.65228419787129e-41 2.85058161779399e-41 3.08721857119471e-41 1.53852388750522e-40 2.66173620735992e-40 4.17021551646017e-40 7.2097712066611e-40 1.34108714945582e-39 2.16569106567114e-39 2.77041615534659e-39 3.38487702676658e-39 0.02585034013605442 0.04217687074829932 0.049913941480206545 0.05505505505505505 0.04014096140080392 0.04556894243641232 0.040922701563712985 0.06021335641724165 0.05178947368421053 0.04583053878828527 0.061016949152542375 0.06471989230609919 0.04197761194029851 0.05131637661758144 0.04395061728395062 0.052725250278086756 0.06677157094151587 0.05772594752186589 0.0600712569392659 0.07013943699026572 0.041445400720258804 0.055421686746987955 0.07104984093319193 0.04732824427480916
1 B B B B B B P B P B P B P B B B B P B P P Bone 580 Colon 33 Gut Lung 558 Midgut 142 Tooth 556 acid mine drainage 117 activated sludge 79 aerobic 392 animal-associated habitat 108 aquaculture 302 aquarium 55 aquatic biome 30 beach sand 191 biofilm 81 bioreactor 166 biotic mesoscopic physical object brine 118 cave 545 coast 17 coastal water 39 compost 425 coral reef 34 fresh water 39 freshwater biome freshwater habitat 12 frost flowers 199 fungi associated P P P P B hot spring 37 hot water 167 hydrothermal vent 313 ice 426 insecta-associated habitat 231 kimchi 16 lake 140 lake sediment 9 lake water 14 large-scale artifact 110 mammalia-associated habitat 105 man-made structure marine biome 16 marine channel 29 marine coral reef biome 50 marine habitat 4 marine reef 80 marine sediment 26 microbial mat 144 mine drainage 566 national park 37 ocean 36 ocean biome 41 oil contaminated soil 141 pasture soil 508 permafrost 289 phyllosphere 375 pond 59 B B rhizosphere soil 0 rumen 17 salt field 68 sea 111 sea floor 80 sea ice 73 sea water 2 seafood product 210 soil 82 solar saltern 131 spring sediment 9 strait 28 stream 167 surface water 4 thermophilic sediment 9 waste water 130 wastewater treatment plant 119 water 47 water column 38 wetland 223 38 P P P P P B P B B B P P P P P B 83 P B 16 P P B B B P P B B B B B Biofilm Ave: 203 Med: 126 Planktonic Ave: 94 Med: 39 P P P P P 140 103
animal associated 582 rock,sand and soil biofilm 584 geographic feature and biome plant associated 377 sediment 572 large-scale artifact 583 Biofilm 550 519 water 583 aerobic environment 394 aquatic feature and biome ice 437 Planktonic 584 waste treatment 562 food 424 artificial natural environment 547 organic feature and biome fungi associated human activity association hydrosphere 584 organism association 584 0 314 584
@prefix xsd: <http://www.w3.org/2001/XMLSchema#> . @prefix msv: <http://purl.jp/bio/11/msv/> . @prefix srs: <http://www.ncbi.nlm.nih.gov/sra/> . @prefix ncbi_refseq: <http://www.ncbi.nlm.nih.gov/nuccore/> . @prefix ko: <http://www.genome.jp/dbget-bin/www_bget?> . @prefix path: <http://www.genome.jp/dbget-bin/www_bget?> . srs:DRS000009 msv:containedKO [ msv:assignedKO ko:K04033; msv:KOCount ]. "2.21645157258498e-05"^^xsd:float; srs:DRS000009 msv:containedKO [ msv:assignedKO ko:K00937; msv:KOCount ]. "2.41630257974925e-05"^^xsd:float; srs:DRS000009 msv:containedKO [ msv:assignedKO ko:K07735; msv:KOCount ]. "1.38171967047323e-05"^^xsd:float;
WilcoxonKO p < 0.05 (336 ) K01610 phosphoenolpyruvatecarboxykinase(ATP) [EC:4.1.1.49] K03761 MFS transporter, MHS family, alpha-ketoglutarate permease K03577 TetR/AcrR family transcriptional regulator, acrAB operon repressor K12538 outer membrane protein HasF K03651 Icc protein K01040 glutaconateCoA-transferase, subunit B [EC:2.8.3.12] K11896 type VI secretion system protein ImpG K03923 modulator of drug activity B K10539 L-arabinose transport system ATP-binding protein [EC:3.6.3.17] K01745 histidine ammonia-lyase[EC:4.3.1.3] K02168 choline/glycine/prolinebetaine transport protein K00322 NAD(P) transhydrogenase[EC:1.6.1.1] K01246 DNA-3-methyladenine glycosylaseI [EC:3.2.2.20] K01426 amidase [EC:3.5.1.4] K11734 aromaticamino acid transportprotein AroP K17215 inositoltransportsystem ATP-binding protein K02403 flagellar transcriptionalactivatorFlhD K16011 mannose-1-phosphate guanylyltransferase/ mannose-6-phosphate isomerase [EC:2.7.7.13 5.3.1.8] K00171 pyruvate ferredoxin oxidoreductase, delta subunit [EC:1.2.7.1] K00355 NAD(P)H dehydrogenase(quinone) [EC:1.6.5.2] K17320 putative aldouronatetransportsystem permease protein K14519 NADP-dependentaldehyde dehydrogenase [EC:1.2.1.4] K17214 inositoltransportsystem permease protein K03119 taurine dioxygenase [EC:1.14.11.17] K08195 MFS transporter, AAHS family, 4-hydroxybenzoate transporter K07098 K03272 D-beta-D-heptose7-phosphate kinase / D-beta-D-heptose1-phosphate adenosyltransferase [EC:2.7.1.167 2.7.7.70] K00849 galactokinase [EC:2.7.1.6] K08224 MFS transporter, YNFM family, putative membrane transport protein K10979 DNA end-binding protein Ku K10117 raffinose/stachyose/melibiosetransport system substrate-binding protein K02108 F-type H+-transporting ATPase subunit a [EC:3.6.3.14] K03704 cold shock protein (beta-ribbon, CspA family) K10237 trehalose/maltose transport system permease protein K10531 L-ornithine N5-oxygenase [EC:1.13.12.-] K10015 histidine transport system permease protein K03773 FKBP-type peptidyl-prolylcis-trans isomerase FklB [EC:5.2.1.8] K10543 D-xylose transport system substrate-binding protein K03736 ethanolamine ammonia-lyasesmall subunit [EC:4.3.1.7] K07407 alpha-galactosidase[EC:3.2.1.22] (1/5)
WilcoxonKOWilcoxonKO Bio Bio Pla Pla Bio Bio Bio Bio Bio Pla Pla
R Genus Biofilm Planktonic Escherichia Escherichia 103 258 121 14 Escherichia = matrix(c(103,258,2,0), ncol = 2 ) Escherichia [Bio] [Pla] [+] 103 258 [-] 121 14 fisher.test(a) Fisher's Exact Test for Count Data data: Escherichia p-value = 2.041e-36 alternative hypothesis: true odds ratio is not equal to 1 95 percent confidence interval: 1.497846 2.569146 sample estimates: odds ratio [(103/258) / (121/14)] 0.046191
Shannon index= -(value/sum)*log2(value/sum) food 5.565130795835259 fungi associated 6.055905711563868 plant associated 6.642378771462215 biofilm 6.950432001122053 aerobic environment 7.681887098090524 ice 8.122683417915658 large-scale artifact 8.377015366930495 artificial natural environment 8.717709878783833 geographic feature and biome 8.880730867862294 waste treatment 9.93970104147625 animal associated 10.319644405818543 sediment 10.385325333127858 rock,sand and soil 11.586005173411566 water 12.313033474340926 aquatic feature and biome 12.373408565130129
DRS000009 MEO_0000437 MEO_0000137 K00937:2.41630257974925e-05 K07735:1.38171967047323e-05 K03117:2. DRS000201 MEO_0000437 MEO_0000137 MEO_0000437 MEO_0000416 MEO_0000437 MEO_0000026 K08590:2. DRS000406 MEO_0000437 MEO_0000718 K02224:3.31285985940223e-06 K07749:1.98771591564134e-05 K00163:1. DRS000407 MEO_0000437 MEO_0000718 K02793:9.26535036922421e-06 K03076:9.26535036922421e-06 K00384:9. ERS000029 MEO_0000437 MEO_0000658 MEO_0000439 MEO_0000439 MEO_0000439 K03086:0.333333333333333 ERS007239 MEO_0000437 MEO_0000056 K07735:0.000704225352112676 K03117:0.00727663895057533 K00954:0. ERS007241 MEO_0000437 MEO_0000056 K01952:0.00181833182080686 K07749:0.000869565217391304 K00163:0. ERS015567 MEO_0000437 MEO_0000024 MEO_0000437 MEO_0000027 MEO_0000437 MEO_0000038 K06999:0. ERS015568 MEO_0000437 MEO_0000038 K02777:0.00294985250737463 K02703:0.0206489675516224 K04077:0. ERS016117 MEO_0000437 MEO_0000449 MEO_0000439 K03117:0.000471119694989063 K00954:0.0087782623731457 ERS016118 MEO_0000437 MEO_0000458 MEO_0000439 K00954:0.000635002877817424 K02224:9.98980764944161e ERS033180 MEO_0000437 MEO_0000704 K00549:0.000175100682892663 K13639:8.75503414463316e-05 K00163:0. SRS000238 MEO_0000437 MEO_0000122 K07084:0.00202839756592292 K01825:0.00101419878296146 K06969:0. SRS000239 MEO_0000437 MEO_0000122 K02275:0.00079428117553614 K03076:0.0039714058776807 K04046:0. SRS000240 MEO_0000437 MEO_0000122 K03616:0.00272108843537415 K04748:0.00136054421768707 K07180:0. SRS000241 MEO_0000437 MEO_0000122 K02224:0.00150037509377344 K01012:0.000750187546886722 K13482:0. SRS000242 MEO_0000437 MEO_0000122 K00163:0.00207900207900208 K06181:0.00207900207900208 K01956:0.
(76) Bone Colon Gut Lung Midgut Tooth acid mine drainage activated sludge aerobic animal-associated habitat aquaculture aquarium aquatic biome beach sand biofilm bioreactor biotic mesoscopic physical object brine cave coast coastal water compost coral reef fresh water freshwater biome freshwater habitat frost flowers fungi associated hot spring hot water KO 4967 3825 3595 211 3703 2602 2175 3486 2087 2819 1057 268 3231 2593 2588 3490 1782 470 1752 6 1870 1988 233 4034 3148 132 742 4270 2489 2724 KO K01467 K03046 K03043 K04077 K02358 K03070 K04043 K03701 K02355 K03086 K01955 K03695 K03544 K09014 K03798 K K03046 K03043 K02355 K02358 K01610 K03737 K02886 K02945 K00239 K03070 K00962 K04043 K04077 K05349 K01955 K K01534 K01995 K05786 K03703 K00981 K03483 K00666 K03046 K06919 K03043 K01999 K06147 K02674 K02358 K03406 K K00351 K03043 K04043 K02016 K06180 K03701 K12257 K03286 K06942 K00243 K03046 K03320 K02952 K07270 K02520 K K03043 K03046 K03893 K02355 K02014 K07119 K03296 K00116 K00383 K01681 K00525 K04753 K00265 K06207 K02111 K K03046 K03043 K03086 K02358 K01955 K02355 K03147 K04077 K00525 K03070 K02274 K03701 K04043 K03544 K02112 K K07496 K02014 K01153 K09822 K07267 K07399 K01533 K07646 K00382 K03043 K02274 K02358 K03046 K01601 K00412 K K03046 K02358 K04077 K03043 K02355 K04043 K02112 K01955 K00962 K01937 K02111 K03701 K03798 K03086 K01915 K K03701 K03046 K03043 K02358 K01955 K00666 K00525 K02470 K03070 K00548 K01869 K02355 K04077 K02112 K00265 K K03046 K03043 K04077 K03696 K07637 K02945 K02274 K00239 K03086 K01681 K02355 K09687 K01955 K03596 K02470 K K08151 K00879 K15269 K03406 K02358 K03046 K03596 K04077 K04043 K01915 K02355 K03296 K02337 K01999 K03043 K K02112 K02689 K01467 K03149 K03438 K00209 K06940 K01915 K02004 K02274 K00658 K06207 K02276 K04096 K03076 K K07637 K07660 K07319 K07496 K01494 K06926 K03465 K07491 K02355 K01190 K02358 K03043 K02112 K03046 K00383 K K03046 K02358 K04077 K03043 K03086 K03070 K01955 K03695 K02112 K03798 K03701 K03782 K02355 K04043 K01681 K K06895 K03545 K01868 K00574 K01951 K13935 K00626 K02575 K07798 K03406 K03070 K00799 K01712 K00426 K06158 K K02358 K03046 K02355 K03737 K03043 K03701 K06147 K01873 K01153 K02004 K03702 K01872 K03070 K03040 K01870 K K06147 K02004 K03657 K03046 K16787 K03701 K01421 K16786 K07024 K03316 K02529 K00383 K01533 K03294 K03763 K K00558 K01955 K03046 K00090 K02111 K01681 K03086 K03406 K02470 K04077 K01873 K03701 K02343 K02621 K00059 K K03046 K03043 K02358 K04077 K03070 K04043 K03086 K02355 K01955 K03696 K03701 K03596 K03695 K02945 K02111 K K00548 K01915 K00342 K04043 K17321 K05559 K03043 K03046 K02014 K02358 K02945 K02355 K03406 K06207 K02111 K11384 K04077 K02886 K04043 K02112 K01955 K K03046 K03043 K02358 K02355 K04077 K02274 K04043 K01915 K03070 K00525 K03701 K01955 K01681 K03702 K09014 K K08884 K02112 K01467 K01711 K06200 K04567 K06160 K02689 K02044 K01638 K02470 K07124 K07237 K09686 K01474 K K00558 K04077 K08151 K04043 K02703 K01711 K03695 K02358 K15269 K01681 K02314 K02040 K03797 K00432 K00525 K K00558 K03111 K02357 K07496 K02835 K01999 K11942 K01653 K02757 K02756 K02755 K02355 K04094 K01966 K06168 K K00558 K03111 K02357 K01999 K07496 K02835 K01653 K11942 K02757 K02755 K02756 K04043 K04094 K02355 K01966 K K03046 K03043 K02358 K00548 K00525 K03296 K01687 K02355 K01955 K00265 K04077 K02198 K01952 K03522 K01915 K K02014 K03296 K00656 K03046 K00123 K01652 K03043 K02761 K00370 K01223 K00688 K00615 K00265 K05802 K00525 K K07637 K07660 K07319 K07496 K01494 K06926 K03465 K07491 K02355 K02358 K03043 K00383 K03046 K02112 K03305 K K03497 K00638 K01190 K00860 K03702 K00833 K01992 K00652 K06890 K03296 K06132 K03147 K01935 K03639 K07347 K
KO K00001 -10.311816354878324 -8.104774922411162 -9.12536274284216 -9.630882031115686 -10.446304307863649 -9.444659766985191 K00002 -13.141335312924086 -16 -12.704600872109006 -14.043777560434304 -12.665170969217156 -10.390757105237586 K00003 -7.121752898407186 -8.413313412355846 -7.382105014212423 -8.475435715319305 -8.852086421482788 -7.445159144381894 K00004 -13.213722194807316 -16 -12.412278655972163 -16 -16 -11.493048389416982 K00005 -9.96363701697749 -8.407649758618964 -12.14236250205763 -10.414517054242882 -16 -11.493048389416982 K00007 -10.596788922173955 -9.329282052339648 -12.918984333818889 -16 -16 -9.409468928010325 K00008 -11.584457072149807 -13.31394247457744 -10.254085944514278 -12.176686333973533 -16 -10.095757399493332 K00009 -11.006367458916358 -8.16761864984201 -12.4902708126153 -11.03250817989423 -16 -11.482051760613226 K00010 -9.264325381071723 -9.186300492329945 -9.45947148340929 -9.3754727463094 -10.469939085997646 -9.88140145587432 K00011 -16 -16 -15.131461775818128 -16 -16 -16 K00012 -7.106372296508517 -6.98777647385517 -7.183819812492088 -7.964181219197697 -6.947145554622075 -6.859389052643327 K00013 -7.7918148582413504 -8.211241559923812 -7.816358527328419 -7.338291834838282 -10.451770159980867 -7.902794931405587 K00014 -9.277120817347368 -8.54307527927404 -8.46073500012144 -8.684299576853675 -10.469939085997646 -8.544543938241562 K00015 -10.73818076223137 -14.628162042310528 -8.362338719924006 -9.334936622441244 -8.002924975011636 -8.778385710633545 K00016 -10.129216802977284 -7.4845566904656415 -10.051986464809646 -11.063919929388838 -16 -11.493048389416982 K00018 -13.225287537775754 -12.25656699684002 -10.968406047051388 -13.672214004001823 -8.843329333909496 -9.183850818741712 K00019 -9.625883141561934 -9.755969817231259 -8.47209897699472 -9.645816479474618 -8.30933106575223 -7.773560661928158 K00020 -9.315047048838318 -9.92986455302942 -8.044325803389565 -9.261050917282379 -9.544127635689426 -7.384846155199226 K00021 -16.245565046960508 -9.061901547084124 -12.748580411992254 -13.246077759947227 -10.446304307863649 -11.482051760613226 K00023 -8.702620832331586 -9.834925987742565 -7.746683551255264 -8.53001839575091 -9.203602442460852 -7.990860797863764 K00024 -6.574629532570801 -7.580959967649561 -5.292461661964257 -7.523288272986434 -7.8223424865027535 -6.844201977115136 K00027 -6.679113848508194 -7.299539418709296 -7.41204210776536 -8.079553675448148 -8.480051144947907 -9.287260449470113 K00028 -16 -13.239973934562654 -16 -16 -16 -16 K00029 -6.88173249202305 -8.052890251183237 -6.470484779211342 -6.858808198174476 -6.8971375085262006 -5.863457907014847 K00030 -9.996294929960587 -9.996499401331711 -9.241982940346352 -8.841324475788843 -10.968721679793427 -9.2945959757397 K00031 -5.604052648376666 -7.0236236350845065 -5.736622818699684 -6.02264419242527 -5.888872580320814 -5.9616659039450965 K00032 -14.554995368110623 -16 -11.621793774892419 -14.554603184200294 -10.968721679793427 -16 K00033 -6.783449648079597 -7.386708390371603 -6.756252671315078 -7.606976602999303 -6.9025269661364295 -7.022195787939136 K00034 -11.068633989656037 -16 -12.067132542569965 -10.063455860550718 -16 -9.08913996971469 K00035 -15.532192564464934 -16 -12.057015551