Correlation Analysis on Colon Lesions and Diseases
Representative immunohistochemistry images of colon lesions in ICRPs with risk alleles and correlation analysis on individuals with non-risk alleles. The study examines the levels of MMP9, phospho-STAT3, phosphor-NF-kB, plasminogen, and their interplay in colons of ICRPs, UC, CD patients, and HC. Significant correlations between these factors are observed, shedding light on potential mechanisms underlying various conditions.
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
You are allowed to download the files provided on this website for personal or commercial use, subject to the condition that they are used lawfully. All files are the property of their respective owners.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.
E N D
Presentation Transcript
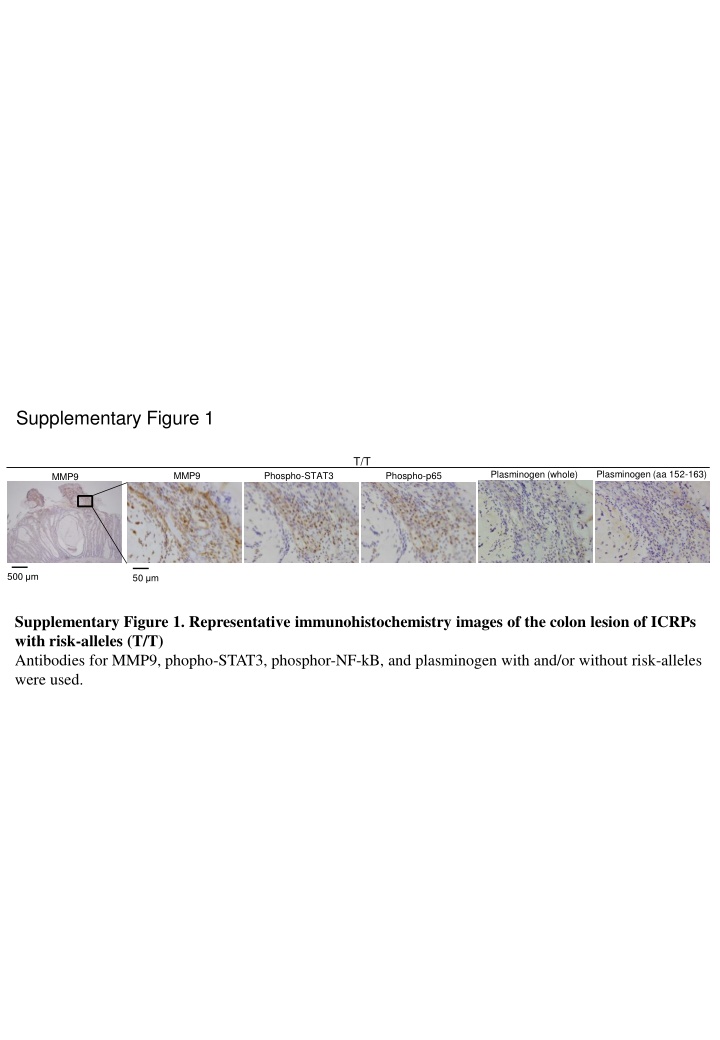
Supplementary Figure 1 T/T Plasminogen (aa 152-163) Plasminogen (whole) MMP9 Phospho-STAT3 Phospho-p65 MMP9 500 m 50 m Supplementary Figure 1. Representative immunohistochemistry images of the colon lesion of ICRPs with risk-alleles (T/T) Antibodies for MMP9, phopho-STAT3, phosphor-NF-kB, and plasminogen with and/or without risk-alleles were used.
Supplementary Figure 2 Supplementary Figure 2. Correlation analysis on the images of the colons of ICRPs with non-risk- alleles (A/A) or risk-alleles (T/T) The signals positive for MMP9, phopho-STAT3, phosphor-NF-kB (phopho-p65), plasminogen (whole), and plasminogen (aa 152-163) at the same lumen structures were quantified. The percentages of the positive areas in the images of the colons of ICRP with non-risk-alleles (open circles) and with risk-alleles (gray triangles) are plotted, and the regression lines are depicted in the figures. The Pearson correlation coefficient (r) and p-value (p) are shown on the right of each figure. The levels of MMP9 were positively correlated with those of phospho-p65 or phospho-STAT3, but the levels of plasminogen (whole) were negatively correlated with those of MMP9, phospho-p65, or phospho-STAT3 in the colons of ICRPs with and without the risk-alleles.
Supplementary Figure 3 Supplementary Figure 3. Correlation analysis on the images of colons of the patients with the human ulcerous colitis (UC) and Crohn s disease (CD), and those of healthy control (HC) The signals positive for MMP9, phopho-STAT3, phosphor-NF-kB (phopho-p65), and plasminogen (whole) at the same lumen structures were quantified. The percentages of the positive areas in the images of the human ulcerous colitis (gray triangles), Crohn s disease (black squares), and healthy control (open circles) are plotted, and the regression lines are depicted in the figures. The Pearson correlation coefficient (r) and p-value (p) are shown on the right of each figure. The levels of MMP9 were positively correlated with those of phospho-p65 or those of phospho-STAT3, but the levels of plasminogen were negatively correlated with those of MMP9 in the colons of the UC and CD patients and HC.
Supplementary Table 1. Signalment and diseases of dogs for whole-exome sequencing. Case number 1 2 3 4 Breed Disease Age 9 9 1 1 Sex Miniature Dachshund Miniature Dachshund Beagle Beagle Inflammatory colorectal polyps Inflammatory colorectal polyps Healthy Healthy Male (neutered) Male Female Female
Supplementary Table 2. Signalment and diseases of dogs for Sanger sequencing. Case number 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 Breed Disease ICRP ICRP ICRP ICRP ICRP ICRP ICRP ICRP ICRP ICRP ICRP ICRP ICRP ICRP ICRP ICRP ICRP ICRP ICRP ICRP ICRP ICRP ICRP ICRP ICRP ICRP ICRP ICRP ICRP ICRP ICRP ICRP ICRP ICRP ICRP ICRP ICRP ICRP ICRP ICRP Age 8 12 11 9 7 6 12 11 10 7 10 13 10 11 12 7 9 8 10 7 Sex Case number 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 Breed Disease Healthy Healthy Healthy Age 13 8 8 5 6 5 4 4 Sex Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Miniature Dachshund Toy Poodle Shiba Shih Tzu Jack Russel Terrier Chihuahua Miniature Schunauzer Cavalier King CharlesSpaniel Welsh Corgi Papillon Labrador Retriever Border Collie Mongrel Mongrel Beagle Beagle Yorkshire Terrier Welsh Corgi Yorkshire Terrier Chihuahua Shih Tzu Wippet Welsh Corgi Labrador Retriever Shih Tzu Toy Poodle Beagle Beagle Beagle Beagle French Bulldog Toy Poodle Italian Greyhound Jack Russel Terrier Chihuahua Mongrel Rottweiler Miniature Schunauzer American Cocker Spaniel Beagle Mongrel Male (neutered) Male (neutered) Female (spayed) Female (spayed) Male (neutered) Male (neutered) Female (spayed) Male (neutered) Male (neutered) Female (spayed) Female (spayed) Male (neutered) Female (spayed) Female (spayed) Female Male Male (neutered) Male (neutered) Intervertebral disk disease Intervertebral disk disease Healthy Healthy Healthy Healthy Healthy Healthy Healthy Healthy Healthy Gallbladder sludge Nasal tumor Epilepsy Healthy Epilepsy Liver nodular hyperplasia Liver tumor Chronic kidney disease Chronic kidney disease Idiopathic vestibular disease Tracheobronchomalacia Intervertebral disk disease Chronic kidney disease Myxomatous mitral vulve disease Interstitial pneumonia Perineal hernia Healthy Anemia Protein-losing enteropathy Pulmorary hypertension Healthy Healthy Acute gastroenteritis Hyperadrenocorticism Chronic enteropathy Myxomatous mitral vulve disease Malassezia dermatitis Intervertebral disk disease Healthy Fibrosarcoma Oral melanoma Colitis Healthy Healthy Healthy Healthy Intervertebral disk disease Pancreatic tumor Lymphoma Idiopathic vestibular disease Plasmacytoma Exocrine pancreatic insufficiency Hypoadrenocorticism Lymphoma Immune-mediated hemolytic anemia Healthy Healthy Male (neutered) 6 6 8 7 13 13 11 15 13 15 16 11 Female (spayed) 14 Female (spayed) 12 Female (spayed) 13 Female (spayed) 11 Female (spayed) 13 Female (spayed) 9 14 Male (neutered) 13 10 Female (spayed) 11 Female 8 Female (spayed) 1 Female 1 Female 13 Male (neutered) 13 Male (neutered) 12 Male (neutered) 11 Female 10 Male (neutered) 12 Female (spayed) 10 Male (neutered) 15 Female 9 Female (spayed) 9 Female (spayed) 1 Female 1 Female 1 Female 1 Female 7 Female (spayed) 11 Female (spayed) 10 Female (spayed) 14 Male (neutered) 14 Female (spayed) 1 Female (spayed) 10 Female (spayed) 13 Female (spayed) 12 Female (spayed) 1 Female 5 Male (neutered) Male (neutered) Male (neutered) Female (spayed) Male (neutered) Female (spayed) Male (neutered) Female (spayed) Male (neutered) Female Female (spayed) Male Male (neutered) Female (spayed) Male Female (spayed) Male (neutered) Male (neutered) Male Male Female (spayed) Male Male Female Female (spayed) Male (neutered) Male Male (neutered) Female (spayed) Female (spayed) 14 7 9 Male 9 14 12 7 11 12 Male Male 7 Male (neutered) 9 9 13 11 11 11 10 8 12 9 12 13 14 9 9 13 12 14 13 4 16 4 7 9 Male Female (spayed) Male (neutered) Male (neutered) Male Male Female (spayed) Male (neutered) Female (spayed) Female (spayed) Female (spayed) Female (spayed) Female (spayed) Male Male Female (spayed) Female (spayed) Female Female (spayed) Female (spayed) Female (spayed) Female (spayed) Female (spayed) Female (spayed) Progressive retinal atrophy Healthy Canine atopic dermatitis Healthy Healthy Healthy Gastroinytestinal stromal tumor Healthy Gallbladder stone Primary hyperparathyroidism Healthy Hyperadrenocorticism Intervertebral disk disease Intervertebral disk disease Healthy Pyometra Healthy Healthy Chronic kidney disease Healthy Healthy Healthy Male
Supplementary Table 3. PCR primers used for Sanger sequencing. Gene NOX3 NOX3 PLG TCOF1 ITGAD CDH2 PTGER2 PTGER2 PTGER2 PTGER2 TSHR GGTA1 ARHGAP8 PDGFB UBE2N DLA-64 TG COL9A2 CYP4A11 RNASEH1 cOR52N9 GIF CD40 IL29L COL4A4 ZDHHC8 ZDHHC8 ZDHHC8 KLRA1 RPL4 FGF5 Variant position 45162067 45185531 49524251 58729174 16822801 60887580 28523355 28523359 28523364 28523406 53480138 61028286 21130864 25817180 41470463 985354 29406010 2649008 13673059 2127184 28969172 50350525 33381986 36440803 39893325 29200529 29200537 29200552 35149078 30767024 4509367 Forward primer (5'-3') CCTGTGTGCTGTGATTGTGC TCACTGCCTGCCTTTGTTGA AGGAGGGAGCGAAATTTGCA GTTCTCAGCCTGTCCCTTCC CCCCAAAAGGAGCATGGCTA AGGTGGATGAAGACGGCATG CCTCCACCATGGGCAGTATC CCTCCACCATGGGCAGTATC CCTCCACCATGGGCAGTATC CCTCCACCATGGGCAGTATC GCTCTCCTGGGCAATGTCTT CTGAAGTTTGGGCTGTGGGA TCAGGCCTCGTCAAAGTGAC GGAAGGTCGTTGGGGTAAGG CTTCCACTGCTCCGCTACAT CAGCATGGTCACAACAGTGC GATGGGCGGTGAAGGGTTAA GCCTTGCGAACGATTGGATC AGATGTGCATGTGTCCAGGG CGCCCGTTCGATTCAATGAC TGGCCCCAGGATTCTTTGTC CCTCCAAAATCTCCTGCCGT CACTCCTCAGTTCTGGCGAG AAAGCAGAGGAGAGTTGGCC TTTGGCACCTTTCACCCCTT CGGCAGGAAGCTGTAGGATC CGGCAGGAAGCTGTAGGATC CGGCAGGAAGCTGTAGGATC CCATCACCTGGGGGAAATGA GCCCTGGAGGAGTGCTAAAT GGCCCTACAAGATGCGCTTA Reverse primer (5'-3') TCTGTTTTCCCCAGCTTCGT ATGTCAGACACCACATGGCA GAGGGGCTGGAGGAGAACTA CGGTGGTGAAGGTCTTGACA CTCCCTCTGCACAGTGGATG GGCTTCCACCAGAGGGAGTA GAACACCAGCTCTGTCACCA GAACACCAGCTCTGTCACCA GAACACCAGCTCTGTCACCA GAACACCAGCTCTGTCACCA CAGGGCAAGAGGTGTCTCAG GCTCTAGCTCTAGGTGCTGC GTTGCCTTTCTTCCTCGGGA CATCCCATGCTCACACACCT TGGCTGCTCCTGTGTTCAAA TCCTCCAGATCACAGCTCCA GCACACAGCCCACAAGAAAG GCTTTAGCCGACACTCAGGT AAGACATGCAGAGGACAGGC AGAAGTTTGCCACGGAGGAG GTGCTGAAGGCTTTGTGACG ACACCCCACTTTCAAGCACA ATCAGGAAACAGGTGCCCAG TTGAACGTGATGGAGGCCTC GTGCAGAAGCTGAGGTCCAT ACCATCACGTATCCTTGGCC ACCATCACGTATCCTTGGCC ACCATCACGTATCCTTGGCC GAACACAGATCCTGGGGTCC GGATGGATAAGGCAGCAGCA GCCATTGACTTTGCCATCCG
Supplementary Table 4. Candidate 31 variants from whole-exome sequencing analysis. Gene NOX3 NOX3 PLG TCOF1 ITGAD CDH2 PTGER2 PTGER2 PTGER2 PTGER2 TSHR GGTA1 ARHGAP8 PDGFB UBE2N DLA-64 TG COL9A2 CYP4A11 RNASEH1 cOR52N9 GIF CD40 IL29L COL4A4 ZDHHC8 ZDHHC8 ZDHHC8 KLRA1 RPL4 FGF5 Chromosome 1 1 1 4 6 7 8 8 8 8 8 9 10 10 10 12 13 15 15 17 21 21 24 24 25 26 26 26 27 30 32 Position 45162067 45185531 49524251 58729174 16822801 60887580 28523355 28523359 28523364 28523406 53480138 61028286 21130864 25817180 41470463 985354 29406010 2649008 13673059 2127184 28969172 50350525 33381986 36440803 39893325 29200529 29200537 29200552 35149078 30767024 4509367 Variant (mRNA) c.967T>C c.242C>T c.478A>T c.349T>C c.1469A>T c.373C>T c.14T>C c.19dupA c.23A>G c.64A>G c.1600C>A c.190T>A c.128G>A c.696C>A c.216T>C c.652A>G c.4567C>T c.88C>T c.1378C>A c.229C>T c.288T>G c.41A>G c.592A>G c.359C>T c.2764G>A c.660A>C c.652A>T c.638G>C c.379C>T c.1226G>C c.284G>T Variant (Protein) p.I323V p.R81K p.N160Y p.S117P p.F490Y p.P125S p.F5S p.N6fs p.Q8R p.T22A p.R534S p.F64I p.P43L p.D232E p.I72M p.I218V p.R1523W p.P30S p.Q460K p.G77S p.F96L p.V14A p.I198V p.T120M p.P922S p.H220Q p.Y218N p.213R p.L127F p.A409G p.C95F MD1 MD2 Variant ID ND ND ND rs24098843 ND ND ND ND ND ND rs9069702 rs24602631 ND ND ND rs22179366 ND ND rs22426403 ND ND rs22919671 rs23176048 rs23189283 ND ND ND ND ND ND ND Variant allele frequency of 10 dogs* 0.95 0.55 0 0 0.6 0.1 1 1 1 1 0.3 0.6 0.2 0.1 0.25 0.4 0 0 0.5 0.4 0.35 0.9 0.75 1 0 1 1 1 0.45 0.05 0.5 Homozygous Heterozygous Heterozygous Homozygous Heterozygous Heterozygous Homozygous Homozygous Homozygous Homozygous Heterozygous Heterozygous Homozygous Homozygous Heterozygous Heterozygous Homozygous Heterozygous Heterozygous Homozygous Homozygous Homozygous Heterozygous Homozygous Heterozygous Homozygous Homozygous Homozygous Homozygous Homozygous Homozygous Homozygous Homozygous Homozygous Homozygous Homozygous Homozygous Homozygous Homozygous Homozygous Homozygous Homozygous Homozygous Homozygous Homozygous Homozygous Homozygous Heterozygous Homozygous Homozygous Homozygous Heterozygous Homozygous Homozygous Homozygous Homozygous Homozygous Homozygous Homozygous Heterozygous Heterozygous Homozygous * Cavalier King Charles Spaniel, Chihuahua, Jack Russell Terrier, Labrador Retriever, Miniature Schnauzer, Papillon, Shiba, Shih Tzu, Toy Poodle, Welsh Corgi