Conditional Statements, Loops, and Functions in R
Learn about conditional statements like if/else, working with loops such as for and while, and creating functions in the R programming language. Understand how to use these fundamental programming concepts to control the flow of your code and perform repetitive tasks efficiently.
Uploaded on Sep 27, 2024 | 0 Views
Download Presentation

Please find below an Image/Link to download the presentation.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.If you encounter any issues during the download, it is possible that the publisher has removed the file from their server.
You are allowed to download the files provided on this website for personal or commercial use, subject to the condition that they are used lawfully. All files are the property of their respective owners.
The content on the website is provided AS IS for your information and personal use only. It may not be sold, licensed, or shared on other websites without obtaining consent from the author.
E N D
Presentation Transcript
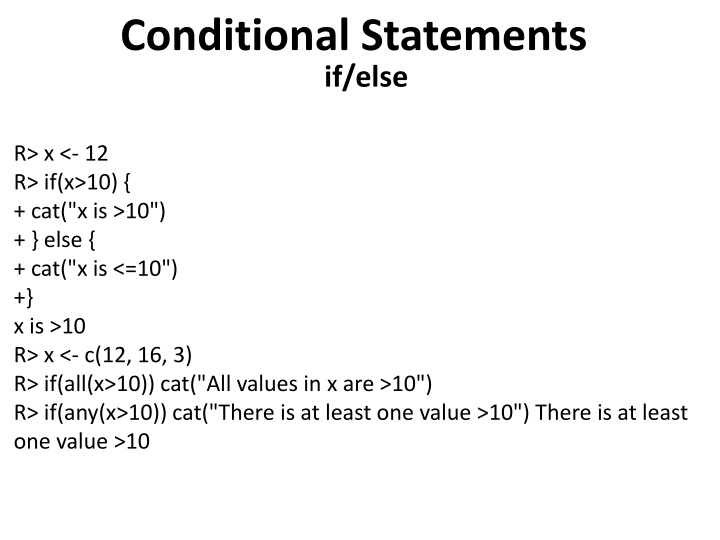
Conditional Statements if/else R> x <- 12 R> if(x>10) { + cat("x is >10") + } else { + cat("x is <=10") +} x is >10 R> x <- c(12, 16, 3) R> if(all(x>10)) cat("All values in x are >10") R> if(any(x>10)) cat("There is at least one value >10") There is at least one value >10
Loops FOR R> x <- 0 R> for(i in 1:5) { + x <- x+i +} R> x [1] 15 R> sum(1:5) [1] 15 R> l <- list(a=2, b=1:2, c=4) R> x <- 0 R> for(i in l) { x <- x+i } R> x [1] 7 8
Loops While R> x <- 0 R> i <- 1 R> while(i <=5) { x <- x+i; i<-i+1 } R> x [1] 15 break and next work as expected. Note: Loops are not very frequently used in R since most problems can be solved more efficiently using functions and vectorization.
Functions R is a functional programming language. Functions are objects of mode function . R> inc <- function(x) { x+1 } R> inc function(x) { x+1 } R> mode(inc) [1] "function" R> inc(5) [1] 6 R> inc(1:10) [1] 2 3 4 5 6 7 8 9 10 11 Since functions are regular (first class) objects they can be passed on as arguments and returned by functions.
Functions An R function is composed by: ? A name that will be used to call the function in the code chunk below, we call our function myFun. ? A set on input formal arguments, that are defined in the parenthesis of the function constructor. The myFun example has two arguments, called i and j. It is possible to provide default values to arguments. ? A function body, with curly brackets (only the body is composed or a single expression). ? A return statement, that represents the output of the function. If no explicit return statement is provided, the last statement of the function is return by default. Functions only support single value. To return multiple values one needs to return a list of the respective return variables like return(list(i, j)). R> myFun <- function(i, j = 1) { + mn <- min(i, j) + mx <- max(i, j) + k <- rnorm(ceiling(i * j)) + return(k[k > mn/mx]) } R> myFun(1.75, 4.45) [1] 1.5953 0.4874 0.7383 R> myFun(1.75) ## j = 1 by default [1] 0.5758
Functions Named arguments and defaults R> inc <- function(x, by = 1) { x + by } R> inc(5) [1] 6 R> inc(1:5, 10) [1] 11 12 13 14 15 R> inc(1:5, by=10) [1] 11 12 13 14 15 R> inc(by=10, x=1:5) [1] 11 12 13 14 15 R> inc(matrix(1:4, nrow=2), 10) [,1] [,2] [1,] 11 13 [2,] 12 14
Functions functions act on copies of their arguments, leaving the original variables intact. R> x <- 1 R> f1 <- function(x) { + x <- x + 10 +x +} R> R> f1(x) [1] 11 R> x ## unchanged [1] 1
Functions Functions have access to the variables defined outside of their body (global variables), while still keeping then unmodified. R> f2 <- function() { + x <- x + 10 +x +} R> R> f2() [1] 11 R> x ## still unchanged [1] 1
Functions Functions can also be written that generate new functions . R> make.power <- function(n) + function(x) x^n +} R> square <- make.power(2) R> cube <- make.power(3) R> square function(x) x^n <environment: 0x2516358> R> get("n", environment(square)) [1] 2 R> square(2) [1] 4 R> get("n", environment(cube)) [1] 3 R> cube(2) [1] 8
Functions The ... arguments When an arbitrary number of arguments is to be passed to a function, one can use the ... special arguments. R> plt <- function(n, ...) { + plot(1:n, ...) } R> par(mfrow = c(1, 2)) R> plt(5, pch = 19, type = "b") R> plt(10, col = rbramp(10), pch = 15)
apply lapply/sapply apply functions to each element in a lists R> l <- list(1:3, 6, 7:3) R> lapply(l, FUN=function(x) { rev(x) }) [[1]] [1] 3 2 1 [[2]] [1] 6 [[3]] [1] 3 4 5 6 7 R> sapply(l, length) [1] 3 1 5
apply apply apply functions to a matrix R> m <- matrix(1:9, nrow=3) R> apply(m, MARGIN=1, sum) [1] 12 15 18 R> apply(m, MARGIN=2, sum) [1] 61524 R> rowSums(m) [1] 12 15 18 R> colSums(m) [1] 61524
apply apply apply functions to a matrix R> m <- matrix(1:9, nrow=3) R> apply(m, MARGIN=1, sum) [1] 12 15 18 R> apply(m, MARGIN=2, sum) [1] 61524 R> rowSums(m) [1] 12 15 18 R> colSums(m) [1] 61524
Measuring execution time We will be using the system.time function to compare for loops with and without initialisation and the apply functions while iterating over the elements of a list of length N = 10^4. We will then compute the mean of each element of the list and multiply it by its length as implemented in function f. R> N <- 10^4 R> ll <- lapply (sample(N), rnorm) R> f <- function(x) {mean(x) * length(x)} R> res1 <- c() R> system.time({ for (i in 1:length(ll)) res1[i] <- f(ll[[i]]) }) R> res2 <- numeric(length(ll)) R> system.time({ for (i in 1:length(ll)) res2[i] <- f(ll[[i]]) }) R> system.time({ res3 <-sapply(ll,f))}
Exercises 1) Create a vector x by x <- runif(100). Write a function with the name avg_gt with two formal arguments: a vector x and a value gt. The functions computes the average of the values greater than gt in x. Write a version with a loop and if and one version without loops and if statements. 1) Write a function small_matrix which computes the smallest value in each column of a given matrix. Create a random 5 5 matrix to test the function.
Exercises 1) Load the obj test_text.Rdata 2) How many characters and elements test has? 3) Create an obj test2 using test and replacing / characters with o in all test elements. 4) Create an obj test2 using test and replacing < characters with in all test elements. 1) Replace the 1 , 2 and 3 character in each element of test2 with AAA
Finding Regex Matches in String Vectors The grep function takes your regex as the first argument, and the input vector as the second argument. If you pass value=FALSE or omit the value parameter then grep returns a new vector with the indexes of the elements in the input vector that could be (partially) matched by the regular expression. If you pass value=TRUE, then grep returns a vector with copies of the actual elements in the input vector that could be (partially) matched. > grep("a+", c("abc", "def", "cba a", "aa"), perl=TRUE, value=FALSE) [1]1 > grep("a+", c("abc", "def", "cba a", "aa"), perl=TRUE, value=TRUE) [1] "abc" "cba a" "aa" 3 4 The grepl function takes the same arguments as the grep function, except for the value argument, which is not supported. grepl returns a logical vector with the same length as the input vector. > grepl("a+", c("abc", "def", "cba a", "aa"), perl=TRUE) [1] TRUE FALSE TRUE TRUE
Exercises 1) Load the obj test_text.Rdata 2) How many elements contain the at least two l ? 3) How many elements contain 4 to 6 numbers? 4) How many elements contain space characters ? 5) How many elements start and end with at least 4 word character?
Exercises 1) Search from NCBI GE dataset cel[Supplementary Files] and download the text file 2) Using the function scan load the file as test_NCBI OBJ. 3) How many elements contain ftp and GSE ? (use length) 4) How many elements contain the world Affy ? 5) Select each element before the world Affy